BLADE-ON-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1
- PMID: 22751300
- PMCID: PMC3583964
- DOI: 10.4161/psb.20599
BLADE-ON-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1
Abstract
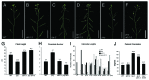
Inflorescence architecture varies widely among flowering plants, serving to optimize the display of flowers for reproductive success. In Arabidopsis thaliana, internode elongation begins at the floral transition, generating a regular spiral arrangement of upwardly-oriented flowers on the primary stem. Post-elongation, differentiation of lignified interfascicular fibers in the stem provides mechanical support. Correct inflorescence patterning requires two interacting homeodomain transcription factors: the KNOTTED1-like protein BREVIPEDICELLUS (BP) and its BEL1-like interaction partner PENNYWISE (PNY). Mutations in BP and PNY cause short internodes, irregular spacing and/or orientation of lateral organs, and altered lignin deposition in stems. Recently, we showed that these defects are caused by the misexpression of lateral organ boundary genes, BLADE-ON-PETIOLE1 (BOP1) and BOP2, which function downstream of BP-PNY in an antagonistic fashion. BOP1/2 gain-of-function in stems promotes expression of the boundary gene KNOTTED1-LIKE FROM ARABIDOPSIS THALIANA6 (KNAT6) and shown here, ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1), providing KNAT6 with a BEL1-like co-factor. Our further analyses show that defects caused by BOP1/2 gain-of-function require both KNAT6 and ATH1. These data reveal how BOP1/2-dependent activation of a boundary module in stems exerts changes in inflorescence architecture.
Figures




Similar articles
-
Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture.Plant Physiol. 2012 Feb;158(2):946-60. doi: 10.1104/pp.111.188573. Epub 2011 Nov 23. Plant Physiol. 2012. PMID: 22114095 Free PMC article.
-
Repression of Lateral Organ Boundary Genes by PENNYWISE and POUND-FOOLISH Is Essential for Meristem Maintenance and Flowering in Arabidopsis.Plant Physiol. 2015 Nov;169(3):2166-86. doi: 10.1104/pp.15.00915. Epub 2015 Sep 28. Plant Physiol. 2015. PMID: 26417006 Free PMC article.
-
ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture.J Exp Bot. 2012 Feb;63(3):1423-33. doi: 10.1093/jxb/err376. Epub 2011 Dec 3. J Exp Bot. 2012. PMID: 22140242 Free PMC article.
-
The Arabidopsis wood model-the case for the inflorescence stem.Plant Sci. 2013 Sep;210:193-205. doi: 10.1016/j.plantsci.2013.05.007. Epub 2013 May 21. Plant Sci. 2013. PMID: 23849126 Review.
-
BLADE-ON-PETIOLE genes: setting boundaries in development and defense.Plant Sci. 2014 Feb;215-216:157-71. doi: 10.1016/j.plantsci.2013.10.019. Epub 2013 Nov 6. Plant Sci. 2014. PMID: 24388527 Review.
Cited by
-
A Gene for Genetic Background in Zea mays: Fine-Mapping enhancer of teosinte branched1.2 to a YABBY Class Transcription Factor.Genetics. 2016 Dec;204(4):1573-1585. doi: 10.1534/genetics.116.194928. Epub 2016 Oct 11. Genetics. 2016. PMID: 27729422 Free PMC article.
-
Nitrogen and Stem Development: A Puzzle Still to Be Solved.Front Plant Sci. 2021 Feb 15;12:630587. doi: 10.3389/fpls.2021.630587. eCollection 2021. Front Plant Sci. 2021. PMID: 33659017 Free PMC article. Review.
-
The Roles of BLH Transcription Factors in Plant Development and Environmental Response.Int J Mol Sci. 2022 Mar 29;23(7):3731. doi: 10.3390/ijms23073731. Int J Mol Sci. 2022. PMID: 35409091 Free PMC article. Review.
-
PHYTOCHROME INTERACTING FACTOR 4 regulates microtubule organization to mediate high temperature-induced hypocotyl elongation in Arabidopsis.Plant Cell. 2023 May 29;35(6):2044-2061. doi: 10.1093/plcell/koad042. Plant Cell. 2023. PMID: 36781395 Free PMC article.
-
OFP1 Interaction with ATH1 Regulates Stem Growth, Flowering Time and Flower Basal Boundary Formation in Arabidopsis.Genes (Basel). 2018 Aug 6;9(8):399. doi: 10.3390/genes9080399. Genes (Basel). 2018. PMID: 30082666 Free PMC article.
References
-
- Wyatt R. Inflorescence architecture: how flower number, arrangement, and phenology affect pollination and fruit set. Am J Bot. 1982;69:585–94. doi: 10.2307/2443068. - DOI
-
- Steeves TA, Sussex IM. Patterns in plant development. 2nd edition. New York: Cambridge University Press, 1989.
-
- Vaughan JG. The morphology and growth of the vegetative and reproductive apices of Arabidopsis thaliana (L.) Heynh., Capsella bursa-pastoris (L.) Medic. and Anagallis arvensis L. Bot J Linn Soc. 1955;55:279–301. doi: 10.1111/j.1095-8339.1955.tb00014.x. - DOI
-
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, et al. Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 2005;42:618–40. doi: 10.1111/j.1365-313X.2005.02403.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
