Comprehensive expression analysis of rice phospholipase D gene family during abiotic stresses and development
- PMID: 22751320
- PMCID: PMC3583975
- DOI: 10.4161/psb.20385
Comprehensive expression analysis of rice phospholipase D gene family during abiotic stresses and development
Abstract
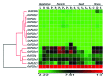
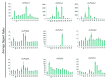
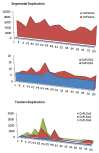
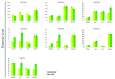
Phospholipase D is one of the crucial enzymes involved in lipid mediated signaling, triggered during various developmental and physiological processes. Different members of PLD gene family have been known to be induced under different abiotic stresses and during developmental processes in various plant species. In this report, we are presenting a detailed microarray based expression analysis and expression profiles of entire set of PLD genes in rice genome, under three abiotic stresses (salt, cold and drought) and different developmental stages (3-vegetative stages and 11-reproductive stages). Seven and nine PLD genes were identified, which were expressed differentially under abiotic stresses and during reproductive developmental stages, respectively. PLD genes, which were expressed significantly under abiotic stresses exhibited an overlapping expression pattern and were also differentially expressed during developmental stages. Moreover, expression pattern for a set of stress induced genes was validated by real time PCR and it supported the microarray expression data. These findings emphasize the role of PLDs in abiotic stress signaling and development in rice. In addition, expression profiling for duplicated PLD genes revealed a functional divergence between the duplicated genes and signify the role of gene duplication in the evolution of this gene family in rice. This expressional study will provide an important platform in future for the functional characterization of PLDs in crop plants.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
