Deletion of the RS domain of RRC1 impairs phytochrome B signaling in Arabidopsis
- PMID: 22751357
- PMCID: PMC3474688
- DOI: 10.4161/psb.20854
Deletion of the RS domain of RRC1 impairs phytochrome B signaling in Arabidopsis
Abstract
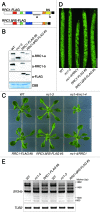
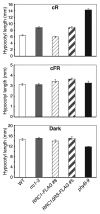
Phytochrome B (phyB), a major photoreceptor in plants, interacts with transcription factors to regulate gene expression and induce various light responses. Recently, we identified an SR-like splicing factor, RRC1 (reduced red-light responses in cry1cry2 background 1), as a novel component of phyB signaling in Arabidopsis. RRC1 has a C-terminal arginine/serine-rich (RS) domain that is generally important for the regulation of alternative splicing. Whereas rrc1 hypomorphic mutant alleles produce truncated RRC1 proteins that lack the C-terminal region, including the RS domain, and exhibit splicing defects and reduced phyB signaling, the rrc1-4 null allele additionally displays pleiotropic developmental abnormalities with more severe splicing defects. Here, we show that transgenic Arabidopsis plants that express truncated RRC1 lacking the RS domain in the rrc1-4 null allele background exhibited the same phenotype as the hypomorphic alleles. Hence, we conclude that deletion of the RS domain of RRC1 reduces phyB signaling, probably due to aberrant regulation of alternative splicing of target genes.
Figures
Similar articles
-
The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction.Plant J. 2012 Jun;70(5):727-38. doi: 10.1111/j.1365-313X.2012.04937.x. Epub 2012 Mar 31. Plant J. 2012. PMID: 22324426
-
Epidermal phyB requires RRC1 to promote light responses by activating the circadian rhythm.New Phytol. 2023 Apr;238(2):705-723. doi: 10.1111/nph.18746. Epub 2023 Feb 13. New Phytol. 2023. PMID: 36651061
-
Coordinated Regulation of Pre-mRNA Splicing by the SFPS-RRC1 Complex to Promote Photomorphogenesis.Plant Cell. 2019 Sep;31(9):2052-2069. doi: 10.1105/tpc.18.00786. Epub 2019 Jul 2. Plant Cell. 2019. PMID: 31266850 Free PMC article.
-
SWAP1-SFPS-RRC1 splicing factor complex modulates pre-mRNA splicing to promote photomorphogenesis in Arabidopsis.Proc Natl Acad Sci U S A. 2022 Nov;119(44):e2214565119. doi: 10.1073/pnas.2214565119. Epub 2022 Oct 25. Proc Natl Acad Sci U S A. 2022. PMID: 36282917 Free PMC article.
-
Integration of light and temperature signaling pathways in plants.J Integr Plant Biol. 2022 Feb;64(2):393-411. doi: 10.1111/jipb.13216. J Integr Plant Biol. 2022. PMID: 34984823 Review.
Cited by
-
Molecular Response of Ulva prolifera to Short-Term High Light Stress Revealed by a Multi-Omics Approach.Biology (Basel). 2022 Oct 25;11(11):1563. doi: 10.3390/biology11111563. Biology (Basel). 2022. PMID: 36358264 Free PMC article.
-
Transcriptional Profiling Reveals the Wheat Defences against Fusarium Head Blight Disease Regulated by a NAC Transcription Factor.Plants (Basel). 2023 Jul 20;12(14):2708. doi: 10.3390/plants12142708. Plants (Basel). 2023. PMID: 37514322 Free PMC article.
-
An improved repertoire of splicing variants and their potential roles in Arabidopsis photomorphogenic development.Genome Biol. 2022 Feb 9;23(1):50. doi: 10.1186/s13059-022-02620-2. Genome Biol. 2022. PMID: 35139889 Free PMC article.
-
HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis.Plant Cell. 2014 Jul;26(7):2858-72. doi: 10.1105/tpc.114.126722. Epub 2014 Jul 22. Plant Cell. 2014. PMID: 25052717 Free PMC article.
-
The Physcomitrella patens chromatin adaptor PpMRG1 interacts with H3K36me3 and regulates light-responsive alternative splicing.Plant Physiol. 2021 Apr 2;185(3):1229-1241. doi: 10.1093/plphys/kiaa103. Plant Physiol. 2021. PMID: 33793927 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
