The subcellular localization of Tubby-like proteins and participation in stress signaling and root colonization by the mutualist Piriformospora indica
- PMID: 22751378
- PMCID: PMC3498949
- DOI: 10.1104/pp.112.201319
The subcellular localization of Tubby-like proteins and participation in stress signaling and root colonization by the mutualist Piriformospora indica
Abstract
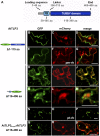
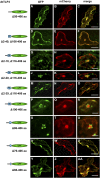
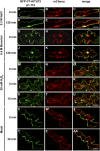
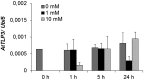
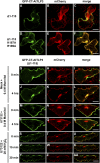
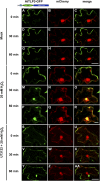
Tubby and Tubby-like proteins (TLPs) were first discovered in mammals, where they are involved in the development and function of neuronal cells. Due to their importance as plasma membrane (PM)-tethered transcription factors or mediators of vesicle trafficking, their lack causes obesity and other disease syndromes. Phosphatidylinositol 4,5-bisphosphate binding of the carboxyl-terminal Tubby domain attaches these proteins to the PM and vesicles and is essential for function. TLPs are conserved across eukaryotic kingdoms including plants, suggesting fundamental biological functions of TLPs. Plant TLPs possess an amino-terminal F-box domain that distinguishes them from other eukaryotic TLPs. Arabidopsis (Arabidopsis thaliana) encodes 11 AtTLPs that fall into six phylogenetic clades. We identified the significance of AtTLPs for root colonization of Arabidopsis by the mutualistic fungus Piriformospora indica. Our results further indicate conserved phosphatidylinositol 4,5-bisphosphate-binding sites in the Tubby domains that are required for PM anchoring of AtTLPs. More detailed studies revealed phospholipase C-triggered release of AtTLP3 from the PM, indicating a conserved mechanism as reported for mammalian Tubby and TLP3. We further show that hydrogen peroxide stimulates the release of AtTLP3 from the PM, presumably for activating downstream events. Different from mammalian homologs, the amino-terminal part of almost all AtTLPs has nucleocytosolic and plastidial localization patterns. Thus, it is tempting to assume that TLPs translate reactive oxygen species currents into signaling not only for transcriptional regulation in the nucleus but also affect plastid-associated functions after release from the PM.
Figures



Similar articles
-
Characterization of Arabidopsis Tubby-like proteins and redundant function of AtTLP3 and AtTLP9 in plant response to ABA and osmotic stress.Plant Mol Biol. 2014 Nov;86(4-5):471-83. doi: 10.1007/s11103-014-0241-6. Epub 2014 Aug 29. Plant Mol Biol. 2014. PMID: 25168737
-
New insights into the subcellular localization of Tubby-like proteins and their participation in the Arabidopsis-Piriformospora indica interaction.Plant Signal Behav. 2013 Aug;8(8):e25198. doi: 10.4161/psb.25198. Epub 2013 Jun 26. Plant Signal Behav. 2013. PMID: 23733076 Free PMC article.
-
Arabidopsis Tubby domain-containing F-box proteins positively regulate immunity by modulating PI4Kβ protein levels.New Phytol. 2023 Oct;240(1):354-371. doi: 10.1111/nph.19187. Epub 2023 Aug 12. New Phytol. 2023. PMID: 37571862 Free PMC article.
-
Tubby-like proteins (TLPs) transcription factor in different regulatory mechanism in plants: a review.Plant Mol Biol. 2022 Dec;110(6):455-468. doi: 10.1007/s11103-022-01301-9. Epub 2022 Oct 18. Plant Mol Biol. 2022. PMID: 36255595 Review.
-
Opprimo ergo sum--evasion and suppression in the root endophytic fungus Piriformospora indica.Mol Plant Microbe Interact. 2012 Jun;25(6):727-37. doi: 10.1094/MPMI-11-11-0291. Mol Plant Microbe Interact. 2012. PMID: 22352718 Review.
Cited by
-
A plant receptor domain with functional analogies to animal malectin disables ER stress responses upon infection.iScience. 2022 Feb 5;25(3):103877. doi: 10.1016/j.isci.2022.103877. eCollection 2022 Mar 18. iScience. 2022. PMID: 35243239 Free PMC article.
-
Attracted to membranes: lipid-binding domains in plants.Plant Physiol. 2021 Apr 2;185(3):707-723. doi: 10.1093/plphys/kiaa100. Plant Physiol. 2021. PMID: 33793907 Free PMC article.
-
Co-immunoprecipitation-based identification of putative BAX INHIBITOR-1-interacting proteins involved in cell death regulation and plant-powdery mildew interactions.Mol Plant Pathol. 2013 Oct;14(8):791-802. doi: 10.1111/mpp.12050. Epub 2013 Jun 19. Mol Plant Pathol. 2013. PMID: 23782494 Free PMC article.
-
Characterization of Arabidopsis Tubby-like proteins and redundant function of AtTLP3 and AtTLP9 in plant response to ABA and osmotic stress.Plant Mol Biol. 2014 Nov;86(4-5):471-83. doi: 10.1007/s11103-014-0241-6. Epub 2014 Aug 29. Plant Mol Biol. 2014. PMID: 25168737
-
A tubby-like protein CsTLP8 acts in the ABA signaling pathway and negatively regulates osmotic stresses tolerance during seed germination.BMC Plant Biol. 2021 Jul 17;21(1):340. doi: 10.1186/s12870-021-03126-y. BMC Plant Biol. 2021. PMID: 34273968 Free PMC article.
References
-
- Abramoff MD, Magalhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 - PubMed
-
- Attard A, Gourgues M, Callemeyn-Torre N, Keller H. (2010) The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol 187: 449–460 - PubMed
-
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci III 316: 1194–1199
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

