PIASy-mediated Tip60 sumoylation regulates p53-induced autophagy
- PMID: 22751435
- PMCID: PMC3409012
- DOI: 10.4161/cc.21091
PIASy-mediated Tip60 sumoylation regulates p53-induced autophagy
Abstract
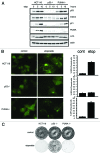
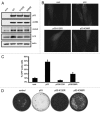
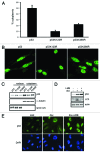
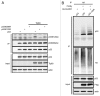
Posttranslational modifications of p53 integrate diverse stress signals and regulate its activity, but their combinatorial contribution to overall p53 function is not clear. We investigated the roles of lysine (K) acetylation and sumoylation on p53 and their relation to apoptosis and autophagy. Here we describe the collaborative role of the SUMO E3 ligase PIASy and the lysine acetyltransferase Tip60 in p53-mediated autophagy. PIASy binding to p53 and PIASy-activated Tip60 lead to K386 sumoylation and K120 acetylation of p53, respectively. Even though these two modifications are not dependent on each other, together they act as a "binary death signal" to promote cytoplasmic accumulation of p53 and execution of PUMA-independent autophagy. PIASy-induced Tip60 sumoylation augments p53 K120 acetylation and apoptosis. In addition to p14(ARF) inactivation, impairment in this intricate signaling may explain why p53 mutations are not found in nearly 50% of malignancies.
Figures


Comment in
-
Directing p53 to induce autophagy.Cell Cycle. 2012 Sep 15;11(18):3353-4. doi: 10.4161/cc.21849. Epub 2012 Aug 23. Cell Cycle. 2012. PMID: 22918242 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
