Deletion of scavenger receptor A gene in mice resulted in protection from septic shock and modulation of TLR4 signaling in isolated peritoneal macrophages
- PMID: 22751446
- PMCID: PMC4349423
- DOI: 10.1177/1753425912449548
Deletion of scavenger receptor A gene in mice resulted in protection from septic shock and modulation of TLR4 signaling in isolated peritoneal macrophages
Abstract
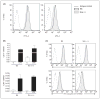
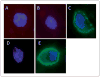
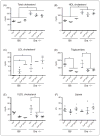
Scavenger receptor A (Sra), also known as macrophage scavenger receptor 1 (Msr1), is a surface glycoprotein preferentially present in macrophages that plays a primary role in innate immunity. Previous studies have shown that Sra is a modifier gene for the response to bacterial LPS in mice at the level of IL-10 production, in particular. In the present study, we found that Sra(-/-) mice are more resistant to septic shock induced by cecal ligation and puncture than wild-type C57BL/6 J (B6) mice. In addition, Sra(-/-) mice displayed initial elevated high density lipoprotein (HDL) circulating levels. Naïve peritoneal macrophages (PMs) were isolated from Sra(-/-) mice to understand the possible protective mechanism. Incubation of these cells with LPS was found to modulate TLR4 signaling, leading to a reduction in IL-10 and IL-6 mRNA levels, but not TNF-α expression, at low concentrations of LPS in comparison with PMs isolated from B6 mice. No differences were found in LPS binding between PMs derived from Sra(-/-) or B6 mice. The lack of Sra binding to LPS was confirmed after transfection of Chinese hamster ovary (CHO) cells with the Sra gene. The contribution of Sra to the outcome of sepsis may be a combination of changes in TLR4 signaling pathway and elevated levels of HDL in circulation, but also LPS toxicity.
Figures




Similar articles
-
Scavenger receptor class a plays a central role in mediating mortality and the development of the pro-inflammatory phenotype in polymicrobial sepsis.PLoS Pathog. 2012;8(10):e1002967. doi: 10.1371/journal.ppat.1002967. Epub 2012 Oct 11. PLoS Pathog. 2012. PMID: 23071440 Free PMC article.
-
Pattern recognition scavenger receptor CD204 attenuates Toll-like receptor 4-induced NF-kappaB activation by directly inhibiting ubiquitination of tumor necrosis factor (TNF) receptor-associated factor 6.J Biol Chem. 2011 May 27;286(21):18795-806. doi: 10.1074/jbc.M111.224345. Epub 2011 Apr 1. J Biol Chem. 2011. PMID: 21460221 Free PMC article.
-
Suppression of TLR4-mediated inflammatory response by macrophage class A scavenger receptor (CD204).Biochem Biophys Res Commun. 2011 Aug 5;411(3):516-22. doi: 10.1016/j.bbrc.2011.06.161. Epub 2011 Jul 2. Biochem Biophys Res Commun. 2011. PMID: 21756882
-
Acute acalculous cholecystitis-like phenotype in scavenger receptor A knock-out mice.J Surg Res. 2012 May 15;174(2):344-51. doi: 10.1016/j.jss.2010.12.033. Epub 2011 Jan 22. J Surg Res. 2012. PMID: 21474146 Free PMC article.
-
The role of macrophage scavenger receptor 1 (MSR1) in inflammatory disorders and cancer.Front Immunol. 2022 Oct 17;13:1012002. doi: 10.3389/fimmu.2022.1012002. eCollection 2022. Front Immunol. 2022. PMID: 36325338 Free PMC article. Review.
Cited by
-
Cysteine-rich domain of scavenger receptor AI modulates the efficacy of surface targeting and mediates oligomeric Aβ internalization.J Biomed Sci. 2013 Aug 2;20(1):54. doi: 10.1186/1423-0127-20-54. J Biomed Sci. 2013. PMID: 23915271 Free PMC article.
-
Scavenger receptor A1 participates in uptake of Leptospira interrogans serovar Autumnalis strain 56606v and inflammation in mouse macrophages.Emerg Microbes Infect. 2021 Dec;10(1):939-953. doi: 10.1080/22221751.2021.1925160. Emerg Microbes Infect. 2021. PMID: 33929941 Free PMC article.
-
Elevated expression of IL-23/IL-17 pathway-related mediators correlates with exacerbation of pulmonary inflammation during polymicrobial sepsis.Shock. 2014 Sep;42(3):246-55. doi: 10.1097/SHK.0000000000000207. Shock. 2014. PMID: 24978886 Free PMC article.
-
Expression of Macrophage Scavenger Receptor (MSR1) in Peripheral Blood Cells from Patients with Different Respiratory Diseases: Beyond Monocytes.J Clin Med. 2022 Mar 5;11(5):1439. doi: 10.3390/jcm11051439. J Clin Med. 2022. PMID: 35268530 Free PMC article.
-
Critical regulation of inflammation via class A scavenger receptor.Int J Chron Obstruct Pulmon Dis. 2018 Apr 13;13:1145-1155. doi: 10.2147/COPD.S153326. eCollection 2018. Int J Chron Obstruct Pulmon Dis. 2018. PMID: 29695898 Free PMC article.
References
-
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Res. 2009;227:221–233. - PubMed
-
- Krieger M, Herz J. Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP) Ann Rev Biochem. 1994;63:601–637. - PubMed
-
- Ashkenas J, Penman M, Vasile E, et al. Structures and high and low affinity ligand binding properties of murine type I and type II macrophage scavenger receptors. J Lipid Res. 1993;34:983–1000. - PubMed
-
- Bowdish DM, Gordon S. Conserved domains of the class A scavenger receptors: evolution and function. Immunol Rev. 2009;227:19–31. - PubMed
-
- Rohrer L, Freeman M, Kodama T, et al. Coiled-coil fibrous domains mediate ligand binding by macrophage scavenger receptor type II. Nature. 1990;343:570–572. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

