Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway
- PMID: 22751496
- PMCID: PMC3403008
- DOI: 10.1101/gad.195248.112
Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway
Abstract
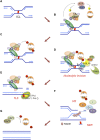
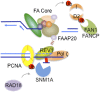
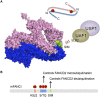
The maintenance of genome stability is critical for survival, and its failure is often associated with tumorigenesis. The Fanconi anemia (FA) pathway is essential for the repair of DNA interstrand cross-links (ICLs), and a germline defect in the pathway results in FA, a cancer predisposition syndrome driven by genome instability. Central to this pathway is the monoubiquitination of FANCD2, which coordinates multiple DNA repair activities required for the resolution of ICLs. Recent studies have demonstrated how the FA pathway coordinates three critical DNA repair processes, including nucleolytic incision, translesion DNA synthesis (TLS), and homologous recombination (HR). Here, we review recent advances in our understanding of the downstream ICL repair steps initiated by ubiquitin-mediated FA pathway activation.
Figures

References
-
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39: 25–35 - PubMed
-
- Alpi AF, Pace PE, Babu MM, Patel KJ 2008. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell 32: 767–777 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
