Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila
- PMID: 22751499
- PMCID: PMC3403011
- DOI: 10.1101/gad.192872.112
Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila
Abstract
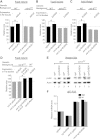
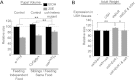
Body size determination is a process that is tightly linked with developmental maturation. Ecdysone, an insect maturation hormone, contributes to this process by antagonizing insulin signaling and thereby suppressing juvenile growth. Here, we report that the microRNA miR-8 and its target, u-shaped (USH), a conserved microRNA/target axis that regulates insulin signaling, are critical for ecdysone-induced body size determination in Drosophila. We found that the miR-8 level is reduced in response to ecdysone, while the USH level is up-regulated reciprocally, and that miR-8 is transcriptionally repressed by ecdysone's early response genes. Furthermore, modulating the miR-8 level correlatively changes the fly body size; either overexpression or deletion of miR-8 abrogates ecdysone-induced growth control. Consistently, perturbation of USH impedes ecdysone's effect on body growth. Thus, miR-8 acts as a molecular rheostat that tunes organismal growth in response to a developmental maturation signal.
Figures



References
-
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T 2003. The small RNA profile during Drosophila melanogaster development. Dev Cell 5: 337–350 - PubMed
-
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell 2: 239–249 - PubMed
-
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 11: 213–221 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
