Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl
- PMID: 22751501
- PMCID: PMC3403015
- DOI: 10.1101/gad.193615.112
Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl
Abstract
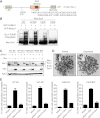
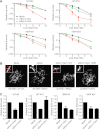
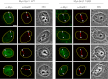
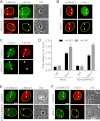
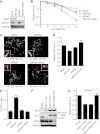
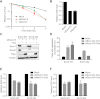
DNA double-strand breaks (DSBs) fuel cancer-driving chromosome translocations. Two related structural maintenance of chromosomes (Smc) complexes, cohesin and Smc5/6, promote DSB repair through sister chromatid homologous recombination (SCR). Here we show that the Smc5/6 subunit Mms21 sumoylates multiple lysines of the cohesin subunit Scc1. Mms21 promotes cohesin-dependent small ubiquitin-like modifier (SUMO) accumulation at laser-induced DNA damage sites in S/G2 human cells. Cells expressing the nonsumoylatable Scc1 mutant (15KR) maintain sister chromatid cohesion during mitosis but are defective in SCR and sensitive to ionizing radiation (IR). Scc1 15KR is recruited to DNA damage sites. Depletion of Wapl, a negative cohesin regulator, rescues SCR defects of Mms21-deficient or Scc1 15KR-expressing cells. Expression of the acetylation-mimicking Smc3 mutant does not bypass the requirement for Mms21 in SCR. We propose that Scc1 sumoylation by Mms21 promotes SCR by antagonizing Wapl at a step after cohesin loading at DSBs and in a way not solely dependent on Smc3 acetylation.
Figures

References
-
- Behlke-Steinert S, Touat-Todeschini L, Skoufias DA, Margolis RL 2009. SMC5 and MMS21 are required for chromosome cohesion and mitotic progression. Cell Cycle 8: 2211–2218 - PubMed
-
- Berkovich E, Monnat RJ Jr, Kastan MB 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 - PubMed
-
- Berkovich E, Monnat RJ Jr, Kastan MB 2008. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc 3: 915–922 - PubMed
-
- De Piccoli G, Torres-Rosell J, Aragon L 2009. The unnamed complex: What do we know about Smc5–Smc6? Chromosome Res 17: 251–263 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
