Synthetic memory circuits for tracking human cell fate
- PMID: 22751502
- PMCID: PMC3403016
- DOI: 10.1101/gad.189035.112
Synthetic memory circuits for tracking human cell fate
Abstract
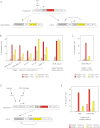
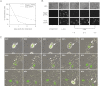
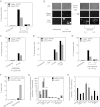
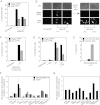
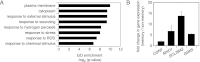
A variety of biological phenomena, from disease progression to stem cell differentiation, are typified by a prolonged cellular response to a transient environmental cue. While biologically relevant, heterogeneity in these long-term responses is difficult to assess at the population level, necessitating the development of biological tools to track cell fate within subpopulations. Here we present a novel synthetic biology approach for identifying and tracking mammalian cell subpopulations. We constructed three genomically integrated circuits that use bistable autoregulatory transcriptional feedback to retain memory of exposure to brief stimuli. These "memory devices" are used to isolate and track the progeny of cells that responded differentially to doxycycline, hypoxia, or DNA-damaging agents. Following hypoxic or ultraviolet radiation exposure, strongly responding cells activate the memory device and exhibit changes in gene expression, growth rates, and viability for multiple generations after the initial stimulus. Taken together, these results indicate that a heritable memory of hypoxia and DNA damage exists in subpopulations that differ in long-term cell behavior.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
