A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development
- PMID: 22751931
- PMCID: PMC3422012
- DOI: 10.1128/MCB.00455-12
A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development
Abstract
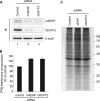
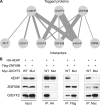
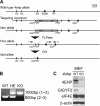
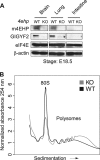
The binding of the eukaryotic initiation factor 4E (eIF4E) to the mRNA 5' cap structure is a rate-limiting step in mRNA translation initiation. eIF4E promotes ribosome recruitment to the mRNA. In Drosophila, the eIF4E homologous protein (d4EHP) forms a complex with binding partners to suppress the translation of distinct mRNAs by competing with eIF4E for binding the 5' cap structure. This repression mechanism is essential for the asymmetric distribution of proteins and normal embryonic development in Drosophila. In contrast, the physiological role of the mammalian 4EHP (m4EHP) was not known. In this study, we have identified the Grb10-interacting GYF protein 2 (GIGYF2) and the zinc finger protein 598 (ZNF598) as components of the m4EHP complex. GIGYF2 directly interacts with m4EHP, and this interaction is required for stabilization of both proteins. Disruption of the m4EHP-GIGYF2 complex leads to increased translation and perinatal lethality in mice. We propose a model by which the m4EHP-GIGYF2 complex represses translation of a subset of mRNAs during embryonic development, as was previously reported for d4EHP.
Figures




Similar articles
-
4EHP-independent repression of endogenous mRNAs by the RNA-binding protein GIGYF2.Nucleic Acids Res. 2018 Jun 20;46(11):5792-5808. doi: 10.1093/nar/gky198. Nucleic Acids Res. 2018. PMID: 29554310 Free PMC article.
-
The eIF4E homolog 4EHP (eIF4E2) regulates hippocampal long-term depression and impacts social behavior.Mol Autism. 2020 Nov 23;11(1):92. doi: 10.1186/s13229-020-00394-7. Mol Autism. 2020. PMID: 33225984 Free PMC article.
-
GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.Genes Dev. 2017 Jun 1;31(11):1147-1161. doi: 10.1101/gad.299420.117. Epub 2017 Jul 11. Genes Dev. 2017. PMID: 28698298 Free PMC article.
-
eIF4E-homologous protein (4EHP): a multifarious cap-binding protein.FEBS J. 2023 Jan;290(2):266-285. doi: 10.1111/febs.16275. Epub 2021 Nov 29. FEBS J. 2023. PMID: 34758096 Review.
-
eIF4E: new family members, new binding partners, new roles.J Biol Chem. 2009 Jun 19;284(25):16711-16715. doi: 10.1074/jbc.R900002200. Epub 2009 Feb 23. J Biol Chem. 2009. PMID: 19237539 Free PMC article. Review.
Cited by
-
Detecting and Rescuing Stalled Ribosomes.Trends Biochem Sci. 2021 Sep;46(9):731-743. doi: 10.1016/j.tibs.2021.03.008. Epub 2021 May 6. Trends Biochem Sci. 2021. PMID: 33966939 Free PMC article. Review.
-
Tracking a refined eIF4E-binding motif reveals Angel1 as a new partner of eIF4E.Nucleic Acids Res. 2013 Sep;41(16):7783-92. doi: 10.1093/nar/gkt569. Epub 2013 Jun 27. Nucleic Acids Res. 2013. PMID: 23814182 Free PMC article.
-
Heterogeneity and specialized functions of translation machinery: from genes to organisms.Nat Rev Genet. 2018 Jul;19(7):431-452. doi: 10.1038/s41576-018-0008-z. Nat Rev Genet. 2018. PMID: 29725087 Free PMC article. Review.
-
4EHP-independent repression of endogenous mRNAs by the RNA-binding protein GIGYF2.Nucleic Acids Res. 2018 Jun 20;46(11):5792-5808. doi: 10.1093/nar/gky198. Nucleic Acids Res. 2018. PMID: 29554310 Free PMC article.
-
eIF4E orchestrates mRNA processing, RNA export and translation to modify specific protein production.Nucleus. 2024 Dec;15(1):2360196. doi: 10.1080/19491034.2024.2360196. Epub 2024 Jun 16. Nucleus. 2024. PMID: 38880976 Free PMC article. Review.
References
-
- Ash MR, et al. 2010. Conserved beta-hairpin recognition by the GYF domains of Smy2 and GIGYF2 in mRNA surveillance and vesicular transport complexes. Structure 18:944–954 - PubMed
-
- Chekulaeva M, Hentze MW, Ephrussi A. 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124:521–533 - PubMed
-
- Cho PF, et al. 2005. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121:411–423 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
