Inactivation of chloramphenicol and florfenicol by a novel chloramphenicol hydrolase
- PMID: 22752166
- PMCID: PMC3416615
- DOI: 10.1128/AEM.01154-12
Inactivation of chloramphenicol and florfenicol by a novel chloramphenicol hydrolase
Abstract
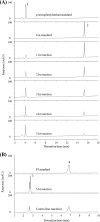
Chloramphenicol and florfenicol are broad-spectrum antibiotics. Although the bacterial resistance mechanisms to these antibiotics have been well documented, hydrolysis of these antibiotics has not been reported in detail. This study reports the hydrolysis of these two antibiotics by a specific hydrolase that is encoded by a gene identified from a soil metagenome. Hydrolysis of chloramphenicol has been recognized in cell extracts of Escherichia coli expressing a chloramphenicol acetate esterase gene, estDL136. A hydrolysate of chloramphenicol was identified as p-nitrophenylserinol by liquid chromatography-mass spectroscopy and proton nuclear magnetic resonance spectroscopy. The hydrolysis of these antibiotics suggested a promiscuous amidase activity of EstDL136. When estDL136 was expressed in E. coli, EstDL136 conferred resistance to both chloramphenicol and florfenicol on E. coli, due to their inactivation. In addition, E. coli carrying estDL136 deactivated florfenicol faster than it deactivated chloramphenicol, suggesting that EstDL136 hydrolyzes florfenicol more efficiently than it hydrolyzes chloramphenicol. The nucleotide sequences flanking estDL136 encode proteins such as amidohydrolase, dehydrogenase/reductase, major facilitator transporter, esterase, and oxidase. The most closely related genes are found in the bacterial family Sphingomonadaceae, which contains many bioremediation-related strains. Whether the gene cluster with estDL136 in E. coli is involved in further chloramphenicol degradation was not clear in this study. While acetyltransferases for chloramphenicol resistance and drug exporters for chloramphenicol or florfenicol resistance are often detected in numerous microbes, this is the first report of enzymatic hydrolysis of florfenicol resulting in inactivation of the antibiotic.
Figures



Similar articles
-
Identification, characterization, and distribution of novel amidase gene aphA in sphingomonads conferring resistance to amphenicol antibiotics.Appl Environ Microbiol. 2024 Nov 20;90(11):e0151224. doi: 10.1128/aem.01512-24. Epub 2024 Oct 21. Appl Environ Microbiol. 2024. PMID: 39431819 Free PMC article.
-
Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea.J Clin Microbiol. 2000 Dec;38(12):4593-8. doi: 10.1128/JCM.38.12.4593-4598.2000. J Clin Microbiol. 2000. PMID: 11101601 Free PMC article.
-
Molecular Mechanism of Chloramphenicol and Thiamphenicol Resistance Mediated by a Novel Oxidase, CmO, in Sphingomonadaceae.Appl Environ Microbiol. 2023 Jan 31;89(1):e0154722. doi: 10.1128/aem.01547-22. Epub 2022 Dec 15. Appl Environ Microbiol. 2023. PMID: 36519886 Free PMC article.
-
Molecular basis of bacterial resistance to chloramphenicol and florfenicol.FEMS Microbiol Rev. 2004 Nov;28(5):519-42. doi: 10.1016/j.femsre.2004.04.001. FEMS Microbiol Rev. 2004. PMID: 15539072 Review.
-
A review on the antibiotic florfenicol: Occurrence, environmental fate, effects, and health risks.Environ Res. 2024 Mar 1;244:117934. doi: 10.1016/j.envres.2023.117934. Epub 2023 Dec 16. Environ Res. 2024. PMID: 38109957 Review.
Cited by
-
Microbial metabolism marvels: a comprehensive review of microbial drug transformation capabilities.Gut Microbes. 2024 Jan-Dec;16(1):2387400. doi: 10.1080/19490976.2024.2387400. Epub 2024 Aug 16. Gut Microbes. 2024. PMID: 39150897 Free PMC article. Review.
-
Discovery and Characterization of a Nitroreductase Capable of Conferring Bacterial Resistance to Chloramphenicol.Cell Chem Biol. 2019 Apr 18;26(4):559-570.e6. doi: 10.1016/j.chembiol.2019.01.007. Epub 2019 Feb 21. Cell Chem Biol. 2019. PMID: 30799223 Free PMC article.
-
History of the streptothricin antibiotics and evidence for the neglect of the streptothricin resistome.NPJ Antimicrob Resist. 2024 Feb 7;2(1):3. doi: 10.1038/s44259-023-00020-5. NPJ Antimicrob Resist. 2024. PMID: 39843956 Free PMC article. Review.
-
Chloramphenicol Derivatives as Antibacterial and Anticancer Agents: Historic Problems and Current Solutions.Antibiotics (Basel). 2016 Jun 3;5(2):20. doi: 10.3390/antibiotics5020020. Antibiotics (Basel). 2016. PMID: 27271676 Free PMC article. Review.
-
Medicinal Chemistry of Inhibitors Targeting Resistant Bacteria.Curr Top Med Chem. 2022;22(24):1983-2028. doi: 10.2174/1568026622666220321124452. Curr Top Med Chem. 2022. PMID: 35319372
References
-
- Arcangioli MA, Leroy-Seètrin S, Martel JL, Chaslus-Dancla E. 1999. A new chloramphenicol and forfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol. Lett. 174: 327–332 - PubMed
-
- Bell SG, Wong LL. 2007. P450 enzymes from the bacterium Novosphingobium aromaticivorans. Biochem. Biophys. Res. Commun. 360: 666–672 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

