Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase
- PMID: 22753026
- PMCID: PMC3458572
- DOI: 10.1093/nar/gks638
Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase
Abstract
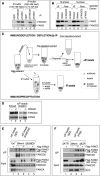
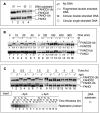
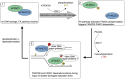
Fanconi anemia (FA) pathway members, FANCD2 and FANCI, contribute to the repair of replication-stalling DNA lesions. FA pathway activation relies on phosphorylation of FANCI by the ataxia telangiectasia and Rad3-related (ATR) kinase, followed by monoubiquitination of FANCD2 and FANCI by the FA core complex. FANCD2 and FANCI are thought to form a functional heterodimer during DNA repair, but it is unclear how dimer formation is regulated or what the functions of the FANCD2-FANCI complex versus the monomeric proteins are. We show that the FANCD2-FANCI complex forms independently of ATR and FA core complex, and represents the inactive form of both proteins. DNA damage-induced FA pathway activation triggers dissociation of FANCD2 from FANCI. Dissociation coincides with FANCD2 monoubiquitination, which significantly precedes monoubiquitination of FANCI; moreover, monoubiquitination responses of FANCD2 and FANCI exhibit distinct DNA substrate specificities. A phosphodead FANCI mutant fails to dissociate from FANCD2, whereas phosphomimetic FANCI cannot interact with FANCD2, indicating that FANCI phosphorylation is the molecular trigger for FANCD2-FANCI dissociation. Following dissociation, FANCD2 binds replicating chromatin prior to-and independently of-FANCI. Moreover, the concentration of chromatin-bound FANCD2 exceeds that of FANCI throughout replication. Our results suggest that FANCD2 and FANCI function separately at consecutive steps during DNA repair in S-phase.
Figures



Similar articles
-
Monoubiquitination by the human Fanconi anemia core complex clamps FANCI:FANCD2 on DNA in filamentous arrays.Elife. 2020 Mar 13;9:e54128. doi: 10.7554/eLife.54128. Elife. 2020. PMID: 32167469 Free PMC article.
-
The DNA-damage kinase ATR activates the FANCD2-FANCI clamp by priming it for ubiquitination.Nat Struct Mol Biol. 2022 Sep;29(9):881-890. doi: 10.1038/s41594-022-00820-9. Epub 2022 Sep 1. Nat Struct Mol Biol. 2022. PMID: 36050501 Free PMC article.
-
Phosphorylation by ATR triggers FANCD2 chromatin loading and activates the Fanconi anemia pathway.Cell Rep. 2023 Jul 25;42(7):112721. doi: 10.1016/j.celrep.2023.112721. Epub 2023 Jun 30. Cell Rep. 2023. PMID: 37392383 Free PMC article.
-
Regulation of the Fanconi Anemia DNA Repair Pathway by Phosphorylation and Monoubiquitination.Genes (Basel). 2021 Nov 5;12(11):1763. doi: 10.3390/genes12111763. Genes (Basel). 2021. PMID: 34828369 Free PMC article. Review.
-
Mechanism, specificity, and function of FANCD2-FANCI ubiquitination and deubiquitination.FEBS J. 2022 Aug;289(16):4811-4829. doi: 10.1111/febs.16077. Epub 2021 Jun 29. FEBS J. 2022. PMID: 34137174 Review.
Cited by
-
FANCI is Associated with Poor Prognosis and Immune Infiltration in Liver Hepatocellular Carcinoma.Int J Med Sci. 2023 May 12;20(7):918-932. doi: 10.7150/ijms.83760. eCollection 2023. Int J Med Sci. 2023. PMID: 37324186 Free PMC article.
-
Fanconi anemia FANCD2 and FANCI proteins regulate the nuclear dynamics of splicing factors.J Cell Biol. 2017 Dec 4;216(12):4007-4026. doi: 10.1083/jcb.201702136. Epub 2017 Oct 13. J Cell Biol. 2017. PMID: 29030393 Free PMC article.
-
Evolution of malignant plasmacytoma cell lines from K14E7 Fancd2-/- mouse long-term bone marrow cultures.Oncotarget. 2016 Oct 18;7(42):68449-68472. doi: 10.18632/oncotarget.12036. Oncotarget. 2016. PMID: 27637088 Free PMC article.
-
Nuclear GIT2 is an ATM substrate and promotes DNA repair.Mol Cell Biol. 2015 Apr;35(7):1081-96. doi: 10.1128/MCB.01432-14. Epub 2015 Jan 20. Mol Cell Biol. 2015. PMID: 25605334 Free PMC article.
-
Head and Neck Cancer Susceptibility and Metabolism in Fanconi Anemia.Cancers (Basel). 2022 Apr 18;14(8):2040. doi: 10.3390/cancers14082040. Cancers (Basel). 2022. PMID: 35454946 Free PMC article. Review.
References
-
- Alter BP, Kupfer G. Fanconi anemia Gene Reviews [Internet] In: Dynan W Dr, editor. Seattle, WA: University of Washington; 1993.
-
- Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2001;2:446–457. - PubMed
-
- Kitao H, Takata M. Fanconi anemia: a disorder defective in the DNA damage response. Int. J. Hematol. 2011;93:417–424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

