RORγ directly regulates the circadian expression of clock genes and downstream targets in vivo
- PMID: 22753030
- PMCID: PMC3458568
- DOI: 10.1093/nar/gks630
RORγ directly regulates the circadian expression of clock genes and downstream targets in vivo
Abstract
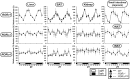
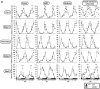
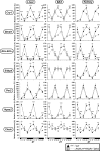
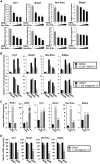
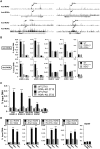
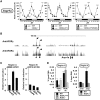
In this study, we demonstrate that the lack of retinoic acid-related orphan receptor (ROR) γ or α expression in mice significantly reduced the peak expression level of Cry1, Bmal1, E4bp4, Rev-Erbα and Per2 in an ROR isotype- and tissue-selective manner without affecting the phase of their rhythmic expression. Analysis of RORγ/RORα double knockout mice indicated that in certain tissues RORγ and RORα exhibited a certain degree of redundancy in regulating clock gene expression. Reporter gene analysis showed that RORγ was able to induce reporter gene activity through the RORE-containing regulatory regions of Cry1, Bmal1, Rev-Erbα and E4bp4. Co-expression of Rev-Erbα or addition of a novel ROR antagonist repressed this activation. ChIP-Seq and ChIP-Quantitative real-time polymerase chain reaction (QPCR) analysis demonstrated that in vivo RORγ regulate these genes directly and in a Zeitgeber time (ZT)-dependent manner through these ROREs. This transcriptional activation by RORs was associated with changes in histone acetylation and chromatin accessibility. The rhythmic expression of RORγ1 by clock proteins may lead to the rhythmic expression of RORγ1 target genes. The presence of RORγ binding sites and its down-regulation in RORγ-/- liver suggest that the rhythmic expression of Avpr1a depends on RORγ consistent with the concept that RORγ1 provides a link between the clock machinery and its regulation of metabolic genes.
Figures



Similar articles
-
Retinoic acid-related orphan receptor γ directly regulates neuronal PAS domain protein 2 transcription in vivo.Nucleic Acids Res. 2011 Jun;39(11):4769-82. doi: 10.1093/nar/gkq1335. Epub 2011 Feb 11. Nucleic Acids Res. 2011. PMID: 21317191 Free PMC article.
-
Prospero-related homeobox 1 (Prox1) functions as a novel modulator of retinoic acid-related orphan receptors α- and γ-mediated transactivation.Nucleic Acids Res. 2013 Aug;41(14):6992-7008. doi: 10.1093/nar/gkt447. Epub 2013 May 30. Nucleic Acids Res. 2013. PMID: 23723244 Free PMC article.
-
Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms.PLoS Genet. 2008 Feb 29;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. PLoS Genet. 2008. PMID: 18454201 Free PMC article.
-
The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes.Prog Nucleic Acid Res Mol Biol. 2001;69:205-47. doi: 10.1016/s0079-6603(01)69048-2. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550795 Review.
-
Dissecting the Rev-erbα Cistrome and the Mechanisms Controlling Circadian Transcription in Liver.Cold Spring Harb Symp Quant Biol. 2015;80:233-8. doi: 10.1101/sqb.2015.80.027508. Epub 2015 Sep 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26370410 Review.
Cited by
-
Circadian clocks and energy metabolism.Cell Mol Life Sci. 2014 Jul;71(14):2667-80. doi: 10.1007/s00018-014-1574-7. Epub 2014 Feb 12. Cell Mol Life Sci. 2014. PMID: 24515123 Free PMC article. Review.
-
Disrupting Hepatocyte Cyp51 from Cholesterol Synthesis Leads to Progressive Liver Injury in the Developing Mouse and Decreases RORC Signalling.Sci Rep. 2017 Jan 18;7:40775. doi: 10.1038/srep40775. Sci Rep. 2017. PMID: 28098217 Free PMC article.
-
Dysregulated circadian rhythm pathway in human osteoarthritis: NR1D1 and BMAL1 suppression alters TGF-β signaling in chondrocytes.Osteoarthritis Cartilage. 2017 Jun;25(6):943-951. doi: 10.1016/j.joca.2016.11.007. Epub 2016 Nov 22. Osteoarthritis Cartilage. 2017. PMID: 27884645 Free PMC article.
-
Vitamin D receptors (VDR), hydroxylases CYP27B1 and CYP24A1 and retinoid-related orphan receptors (ROR) level in human uveal tract and ocular melanoma with different melanization levels.Sci Rep. 2019 Jun 24;9(1):9142. doi: 10.1038/s41598-019-45161-8. Sci Rep. 2019. PMID: 31235702 Free PMC article.
-
GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock.Science. 2015 Jun 26;348(6242):1488-92. doi: 10.1126/science.aab3021. Epub 2015 Jun 4. Science. 2015. PMID: 26044300 Free PMC article.
References
-
- Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol. Genomics. 2007;31:281–294. - PubMed
-
- Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem. 2008;283:18411–18421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

