Transcriptional repression mediated by a TetR family protein, PfmR, from Thermus thermophilus HB8
- PMID: 22753056
- PMCID: PMC3415517
- DOI: 10.1128/JB.00668-12
Transcriptional repression mediated by a TetR family protein, PfmR, from Thermus thermophilus HB8
Abstract
PfmR is one of four TetR family transcriptional regulators found in the extremely thermophilic bacterium, Thermus thermophilus HB8. We identified three promoters with strong negative regulation by PfmR, both in vivo and in vitro. PfmR binds pseudopalindromic sequences, with the consensus sequence of 5'-TACCGACCGNTNGGTN-3' surrounding the promoters. According to the amino acid sequence and three-dimensional structure analyses of the PfmR-regulated gene products, they are predicted to be involved in phenylacetic acid and fatty acid metabolism. In vitro analyses revealed that PfmR weakly cross-regulated with the TetR family repressor T. thermophilus PaaR, which controls the expression of the paa gene cluster putatively involved in phenylacetic acid degradation but not with another functionally identified TetR family repressor, T. thermophilus FadR, which is involved in fatty acid degradation. The X-ray crystal structure of the N-terminal DNA-binding domain of PfmR and the nucleotide sequence of the predicted PfmR-binding site are quite similar to those of the TetR family repressor QacR from Staphylococcus aureus. Similar to QacR, two PfmR dimers bound per target DNA. The bases recognized by QacR within the QacR-binding site are conserved in the predicted PfmR-binding site, and they were important for PfmR to recognize the binding site and properly assemble on it. The center of the PfmR molecule contains a tunnel-like pocket, which may be the ligand-binding site of this regulator.
Figures



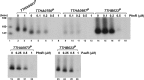


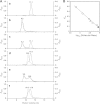
References
-
- Agari Y, Agari K, Sakamoto K, Kuramitsu S, Shinkai A. 2011. TetR family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation. Microbiology 157:1589–1601 - PubMed
-
- Agari Y, Kashihara A, Yokoyama S, Kuramitsu S, Shinkai A. 2008. Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator. Mol. Microbiol. 70:60–75 - PubMed
-
- Brünger AT, et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 - PubMed
-
- Collaborative Computational Project Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

