A major facilitator superfamily protein, HepP, is involved in formation of the heterocyst envelope polysaccharide in the cyanobacterium Anabaena sp. strain PCC 7120
- PMID: 22753066
- PMCID: PMC3415476
- DOI: 10.1128/JB.00489-12
A major facilitator superfamily protein, HepP, is involved in formation of the heterocyst envelope polysaccharide in the cyanobacterium Anabaena sp. strain PCC 7120
Abstract
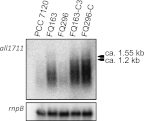
Some filamentous cyanobacteria such as Anabaena sp. strain PCC 7120 produce cells, termed heterocysts, specialized in nitrogen fixation. Heterocysts bear a thick envelope containing an inner layer of glycolipids and an outer layer of polysaccharide that restrict the diffusion of air (including O(2)) into the heterocyst. Anabaena sp. mutants impaired in production of either of those layers show a Fox(-) phenotype (requiring fixed nitrogen for growth under oxic conditions). We have characterized a set of transposon-induced Fox(-) mutants in which transposon Tn5-1063 was inserted into the Anabaena sp. chromosome open reading frame all1711 which encodes a predicted membrane protein that belongs to the major facilitator superfamily (MFS). These mutants showed higher nitrogenase activities under anoxic than under oxic conditions and altered sucrose uptake. Electron microscopy and alcian blue staining showed a lack of the heterocyst envelope polysaccharide (Hep) layer. Northern blot and primer extension analyses showed that, in a manner dependent on the nitrogen-control transcription factor NtcA, all1711 was strongly induced after nitrogen step-down. Confocal microscopy of an Anabaena sp. strain producing an All1711-green fluorescent protein (All1711-GFP) fusion protein showed induction in all cells of the filament but at higher levels in differentiating heterocysts. All1711-GFP was located in the periphery of the cells, consistent with All1711 being a cytoplasmic membrane protein. Expression of all1711 from the P(glnA) promoter in a multicopy plasmid led to production of a presumptive exopolysaccharide by vegetative cells. These results suggest that All1711, which we denote HepP, is involved in transport of glycoside(s), with a specific physiological role in production of Hep.
Figures







Similar articles
-
Specific Glucoside Transporters Influence Septal Structure and Function in the Filamentous, Heterocyst-Forming Cyanobacterium Anabaena sp. Strain PCC 7120.J Bacteriol. 2017 Mar 14;199(7):e00876-16. doi: 10.1128/JB.00876-16. Print 2017 Apr 1. J Bacteriol. 2017. PMID: 28096449 Free PMC article.
-
Alr5068, a Low-Molecular-Weight protein tyrosine phosphatase, is involved in formation of the heterocysts polysaccharide layer in the cyanobacterium Anabaena sp. PCC 7120.Res Microbiol. 2013 Oct;164(8):875-85. doi: 10.1016/j.resmic.2013.05.003. Epub 2013 Jul 1. Res Microbiol. 2013. PMID: 23827083
-
Septum-localized protein required for filament integrity and diazotrophy in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120.J Bacteriol. 2007 May;189(10):3884-90. doi: 10.1128/JB.00085-07. Epub 2007 Mar 16. J Bacteriol. 2007. PMID: 17369306 Free PMC article.
-
The cell wall in heterocyst formation by Anabaena sp. PCC 7120.J Basic Microbiol. 2009 Feb;49(1):5-24. doi: 10.1002/jobm.200800300. J Basic Microbiol. 2009. PMID: 19253332 Review.
-
The Making of a Heterocyst in Cyanobacteria.Annu Rev Microbiol. 2022 Sep 8;76:597-618. doi: 10.1146/annurev-micro-041320-093442. Epub 2022 Jun 7. Annu Rev Microbiol. 2022. PMID: 35671534 Review.
Cited by
-
Inactivation of agmatinase expressed in vegetative cells alters arginine catabolism and prevents diazotrophic growth in the heterocyst-forming cyanobacterium Anabaena.Microbiologyopen. 2014 Oct;3(5):777-92. doi: 10.1002/mbo3.207. Epub 2014 Sep 10. Microbiologyopen. 2014. PMID: 25209059 Free PMC article.
-
Evaluation of self-sustaining cyanobacterial biofilms for technical applications.Biofilm. 2022 Mar 31;4:100073. doi: 10.1016/j.bioflm.2022.100073. eCollection 2022 Dec. Biofilm. 2022. PMID: 35434604 Free PMC article.
-
Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium.Proc Natl Acad Sci U S A. 2014 Mar 11;111(10):3823-8. doi: 10.1073/pnas.1318564111. Epub 2014 Feb 18. Proc Natl Acad Sci U S A. 2014. PMID: 24550502 Free PMC article.
-
Specific Glucoside Transporters Influence Septal Structure and Function in the Filamentous, Heterocyst-Forming Cyanobacterium Anabaena sp. Strain PCC 7120.J Bacteriol. 2017 Mar 14;199(7):e00876-16. doi: 10.1128/JB.00876-16. Print 2017 Apr 1. J Bacteriol. 2017. PMID: 28096449 Free PMC article.
References
-
- Ausubel FM, et al. 2012. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, NY
-
- Awai K, Wolk CP. 2007. Identification of the glycosyl transferase required for synthesis of the principal glycolipid characteristic of heterocysts of Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 266:98–102 - PubMed
-
- Awai K, Lechno-Yossef S, Wolk CP. 2009. Heterocyst envelope glycolipids, p 179–202 In Wada H, Murata N. (ed), Lipids in photosynthesis: essential and regulatory functions. Springer, Heidelberg, Germany
-
- Black TA, Cai Y, Wolk CP. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77–84 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

