Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources
- PMID: 22753413
- PMCID: PMC3436554
- DOI: 10.1074/jbc.M112.372680
Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources
Abstract
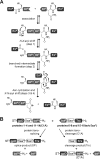
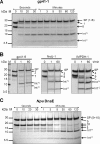
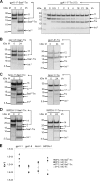
Inteins excise themselves out of precursor proteins by the protein splicing reaction and have emerged as valuable protein engineering tools in numerous and diverse biotechnological applications. Split inteins have recently attracted particular interest because of the opportunities associated with generating a protein from two separate polypeptides and with trans-cleavage applications made possible by split intein mutants. However, natural split inteins are rare and differ greatly in their usefulness with regard to the achievable rates and yields. Here we report the first functional characterization of new split inteins previously identified by bioinformatics from metagenomic sources. The N- and C-terminal fragments of the four inteins gp41-1, gp41-8, NrdJ-1, and IMPDH-1 were prepared as fusion constructs with model proteins. Upon incubation of complementary pairs, we observed trans-splicing reactions with unprecedented rates and yields for all four inteins. Furthermore, no side reactions were detectable, and the precursor constructs were consumed virtually quantitatively. The rate for the gp41-1 intein, the most active intein on all accounts, was k = 1.8 ± 0.5 × 10(-1) s(-1), which is ∼10-fold faster than the rate reported for the Npu DnaE intein and gives rise to completed reactions within 20-30 s. No cross-reactivity in exogenous combinations was observed. Using C1A mutants, all inteins were efficient in the C-terminal cleavage reaction, albeit at lower rates. C-terminal cleavage could be performed under a wide range of reaction conditions and also in the absence of native extein residues flanking the intein. Thus, these inteins hold great potential for splicing and cleavage applications.
Figures

References
-
- Hirata R., Ohsumk Y., Nakano A., Kawasaki H., Suzuki K., Anraku Y. (1990) Molecular structure of a gene, VMA1, encoding the catalytic subunit of H+-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265, 6726–6733 - PubMed
-
- Kane P. M., Yamashiro C. T., Wolczyk D. F., Neff N., Goebl M., Stevens T. H. (1990) Protein splicing converts the yeast TFP1 gene product to the 69-kDa subunit of the vacuolar H+-adenosine triphosphatase. Science 250, 651–657 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

