Amino-terminal cysteine residues differentially influence RGS4 protein plasma membrane targeting, intracellular trafficking, and function
- PMID: 22753418
- PMCID: PMC3436580
- DOI: 10.1074/jbc.M112.345629
Amino-terminal cysteine residues differentially influence RGS4 protein plasma membrane targeting, intracellular trafficking, and function
Abstract
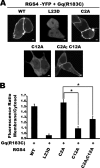
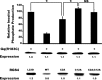
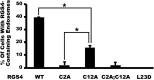
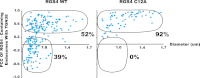
Regulator of G-protein signaling (RGS) proteins are potent inhibitors of heterotrimeric G-protein signaling. RGS4 attenuates G-protein activity in several tissues. Previous work demonstrated that cysteine palmitoylation on residues in the amino-terminal (Cys-2 and Cys-12) and core domains (Cys-95) of RGS4 is important for protein stability, plasma membrane targeting, and GTPase activating function. To date Cys-2 has been the priority target for RGS4 regulation by palmitoylation based on its putative role in stabilizing the RGS4 protein. Here, we investigate differences in the contribution of Cys-2 and Cys-12 to the intracellular localization and function of RGS4. Inhibition of RGS4 palmitoylation with 2-bromopalmitate dramatically reduced its localization to the plasma membrane. Similarly, mutation of the RGS4 amphipathic helix (L23D) prevented membrane localization and its G(q) inhibitory function. Together, these data suggest that both RGS4 palmitoylation and the amphipathic helix domain are required for optimal plasma membrane targeting and function of RGS4. Mutation of Cys-12 decreased RGS4 membrane targeting to a similar extent as 2-bromopalmitate, resulting in complete loss of its G(q) inhibitory function. Mutation of Cys-2 did not impair plasma membrane targeting but did partially impair its function as a G(q) inhibitor. Comparison of the endosomal distribution pattern of wild type and mutant RGS4 proteins with TGN38 indicated that palmitoylation of these two cysteines contributes differentially to the intracellular trafficking of RGS4. These data show for the first time that Cys-2 and Cys-12 play markedly different roles in the regulation of RGS4 membrane localization, intracellular trafficking, and G(q) inhibitory function via mechanisms that are unrelated to RGS4 protein stabilization.
Figures




References
-
- Ross E. M., Wilkie T. M. (2000) GTPase-activating proteins for heterotrimeric G proteins. Regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 - PubMed
-
- Berman D. M., Wilkie T. M., Gilman A. G. (1996) GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein α subunits. Cell 86, 445–452 - PubMed
-
- Watson N., Linder M. E., Druey K. M., Kehrl J. H., Blumer K. J. (1996) RGS family members. GTPase-activating proteins for heterotrimeric G-protein α-subunits. Nature 383, 172–175 - PubMed
-
- Pacey L. K., Heximer S. P., Hampson D. R. (2009) Increased GABA(B) receptor-mediated signaling reduces the susceptibility of fragile X knockout mice to audiogenic seizures. Mol. Pharmacol. 76, 18–24 - PubMed
-
- Cifelli C., Rose R. A., Zhang H., Voigtlaender-Bolz J., Bolz S. S., Backx P. H., Heximer S. P. (2008) RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node. Circ. Res. 103, 527–535 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

