Reshaping of the maize transcriptome by domestication
- PMID: 22753482
- PMCID: PMC3406829
- DOI: 10.1073/pnas.1201961109
Reshaping of the maize transcriptome by domestication
Abstract
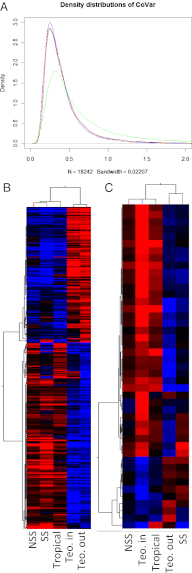
Through domestication, humans have substantially altered the morphology of Zea mays ssp. parviglumis (teosinte) into the currently recognizable maize. This system serves as a model for studying adaptation, genome evolution, and the genetics and evolution of complex traits. To examine how domestication has reshaped the transcriptome of maize seedlings, we used expression profiling of 18,242 genes for 38 diverse maize genotypes and 24 teosinte genotypes. We detected evidence for more than 600 genes having significantly different expression levels in maize compared with teosinte. Moreover, more than 1,100 genes showed significantly altered coexpression profiles, reflective of substantial rewiring of the transcriptome since domestication. The genes with altered expression show a significant enrichment for genes previously identified through population genetic analyses as likely targets of selection during maize domestication and improvement; 46 genes previously identified as putative targets of selection also exhibit altered expression levels and coexpression relationships. We also identified 45 genes with altered, primarily higher, expression in inbred relative to outcrossed teosinte. These genes are enriched for functions related to biotic stress and may reflect responses to the effects of inbreeding. This study not only documents alterations in the maize transcriptome following domestication, identifying several genes that may have contributed to the evolution of maize, but highlights the complementary information that can be gained by combining gene expression with population genetic analyses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Doebley J. The genetics of maize evolution. Annu Rev Genet. 2004;38:37–59. - PubMed
-
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. - PubMed
-
- Freeling M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

