Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites
- PMID: 22753506
- PMCID: PMC3406870
- DOI: 10.1073/pnas.1209309109
Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites
Abstract
Cryptic allosteric sites--transient pockets in a folded protein that are invisible to conventional experiments but can alter enzymatic activity via allosteric communication with the active site--are a promising opportunity for facilitating drug design by greatly expanding the repertoire of available drug targets. Unfortunately, identifying these sites is difficult, typically requiring resource-intensive screening of large libraries of small molecules. Here, we demonstrate that Markov state models built from extensive computer simulations (totaling hundreds of microseconds of dynamics) can identify prospective cryptic sites from the equilibrium fluctuations of three medically relevant proteins--β-lactamase, interleukin-2, and RNase H--even in the absence of any ligand. As in previous studies, our methods reveal a surprising variety of conformations--including bound-like configurations--that implies a role for conformational selection in ligand binding. Moreover, our analyses lead to a number of unique insights. First, direct comparison of simulations with and without the ligand reveals that there is still an important role for an induced fit during ligand binding to cryptic sites and suggests new conformations for docking. Second, correlations between amino acid sidechains can convey allosteric signals even in the absence of substantial backbone motions. Most importantly, our extensive sampling reveals a multitude of potential cryptic sites--consisting of transient pockets coupled to the active site--even in a single protein. Based on these observations, we propose that cryptic allosteric sites may be even more ubiquitous than previously thought and that our methods should be a valuable means of guiding the search for such sites.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
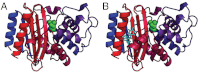

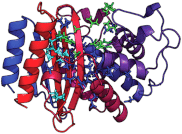

References
-
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. - PubMed
-
- Horn JR, Shoichet BK. Allosteric inhibition through core disruption. J Mol Biol. 2004;336:1283–1291. - PubMed
-
- Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Curr Opin Struct Biol. 2004;14:706–715. - PubMed
-
- Ceccarelli DF, et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–1087. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

