Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers
- PMID: 22753589
- PMCID: PMC3624899
- DOI: 10.1158/1078-0432.CCR-11-3210
Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers
Abstract
Purpose: To assess prognostic roles of various KRAS oncogene mutations in colorectal cancer, BRAF mutation status must be controlled for because BRAF mutation is associated with poor prognosis, and almost all BRAF mutants are present among KRAS wild-type tumors. Taking into account experimental data supporting a greater oncogenic effect of codon 12 mutations compared with codon 13 mutations, we hypothesized that KRAS codon 12-mutated colorectal cancers might behave more aggressively than KRAS wild-type tumors and codon 13 mutants.
Experimental design: Using molecular pathological epidemiology database of 1,261 rectal and colon cancers, we examined clinical outcome and tumor biomarkers of KRAS codon 12 and 13 mutations in 1,075 BRAF wild-type cancers (i.e., controlling for BRAF status). Cox proportional hazards model was used to compute mortality HR, adjusting for potential confounders, including stage, PIK3CA mutations, microsatellite instability, CpG island methylator phenotype, and LINE-1 methylation.
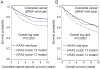
Results: Compared with patients with KRAS wild-type/BRAF wild-type cancers (N = 635), those with KRAS codon 12 mutations (N = 332) experienced significantly higher colorectal cancer-specific mortality [log-rank P = 0.0001; multivariate HR, 1.30; 95% confidence interval (CI), 1.02-1.67; P = 0.037], whereas KRAS codon 13-mutated cases (N = 108) were not significantly associated with prognosis. Among the seven most common KRAS mutations, c.35G>T (p.G12V; N = 93) was associated with significantly higher colorectal cancer-specific mortality (log-rank P = 0.0007; multivariate HR, 2.00; 95% CI, 1.38-2.90, P = 0.0003) compared with KRAS wild-type/BRAF wild-type cases.
Conclusions: KRAS codon 12 mutations (in particular, c.35G>T), but not codon 13 mutations, are associated with inferior survival in BRAF wild-type colorectal cancer. Our data highlight the importance of accurate molecular characterization in colorectal cancer.
©2012 AACR.
Conflict of interest statement
Conflict of interest: The authors have declared no conflicts of interest.
Figures

References
-
- Roock WD, Vriendt VD, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603. - PubMed
-
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–6. - PubMed
-
- Jimeno A, Messersmith WA, Hirsch FR, Franklin WA, Eckhardt SG. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: practical application of patient selection. J Clin Oncol. 2009;27:1130–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

