Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae
- PMID: 22753783
- PMCID: PMC3404376
- DOI: 10.1261/rna.033779.112
Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae
Abstract
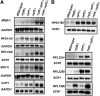
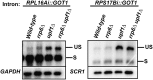
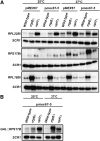
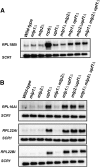
The nuclear exosome and the nonsense-mediated mRNA decay (NMD) pathways have been implicated in the degradation of distinct unspliced transcripts in Saccharomyces cerevisiae. In this study we show that these two systems can act sequentially on specific unspliced pre-mRNAs to limit their accumulation. Using steady-state and decay analyses, we show that while specific unspliced transcripts rely mostly on NMD or on the nuclear exosome for their degradation, some unspliced RNAs are stabilized only when both the nuclear exosome and NMD are inactivated. We found that the mechanism of degradation of these unspliced pre-mRNAs is not influenced by promoter identity. However, the specificity in the pre-mRNAs degradation pathways can be manipulated by changing the rate of export or retention of these mRNAs. For instance, reducing the nuclear export of pre-mRNAs mostly degraded by NMD results in a higher fraction of unspliced transcripts degraded by the nuclear exosome. Reciprocally, inactivating the Mlp retention factors results in a higher fraction of unspliced transcripts degraded by NMD for precursors normally targeted by the nuclear exosome. Overall, these results demonstrate that a functional redundancy exists between nuclear and cytoplasmic degradation pathways for unspliced pre-mRNAs, and suggest that the degradation routes of these species are mainly determined by the efficiency of their nuclear export rates. The presence of these two sequential degradation pathways for unspliced pre-mRNAs underscores the importance of limiting their accumulation and might serve as a fail-safe mechanism to prevent the expression of these nonfunctional RNAs.
Figures

References
-
- Bousquet-Antonelli C, Presutti C, Tollervey D 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 - PubMed
-
- Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483 - PubMed
-
- Chang YF, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
