Transcription factor zinc finger and BTB domain 1 is essential for lymphocyte development
- PMID: 22753936
- PMCID: PMC3401355
- DOI: 10.4049/jimmunol.1200623
Transcription factor zinc finger and BTB domain 1 is essential for lymphocyte development
Abstract
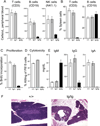
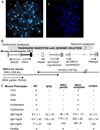
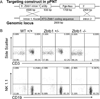
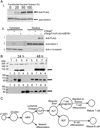
Absent T lymphocytes were unexpectedly found in homozygotes of a transgenic mouse from an unrelated project. T cell development did not progress beyond double-negative stage 1 thymocytes, resulting in a hypocellular, vestigial thymus. B cells were present, but NK cell number and B cell isotype switching were reduced. Transplantation of wild-type hematopoietic cells corrected the defect, which was traced to a deletion involving five contiguous genes at the transgene insertion site on chromosome 12C3. Complementation using bacterial artificial chromosome transgenesis implicated zinc finger BTB-POZ domain protein 1 (Zbtb1) in the immunodeficiency, confirming its role in T cell development and suggesting involvement in B and NK cell differentiation. Targeted disruption of Zbtb1 recapitulated the T(-)B(+)NK(-) SCID phenotype of the original transgenic animal. Knockouts for Zbtb1 had expanded populations of bone marrow hematopoietic stem cells and also multipotent and early lymphoid lineages, suggesting a differentiation bottleneck for common lymphoid progenitors. Expression of mRNA encoding Zbtb1, a predicted transcription repressor, was greatest in hematopoietic stem cells, thymocytes, and pre-B cells, highlighting its essential role in lymphoid development.
Figures



Similar articles
-
Gene coexpression analysis in single cells indicates lymphomyeloid copriming in short-term hematopoietic stem cells and multipotent progenitors.J Immunol. 2010 May 1;184(9):4907-17. doi: 10.4049/jimmunol.0902184. Epub 2010 Apr 5. J Immunol. 2010. PMID: 20368277
-
Mobilization of human lymphoid progenitors after treatment with granulocyte colony-stimulating factor.J Immunol. 2005 Aug 15;175(4):2647-54. doi: 10.4049/jimmunol.175.4.2647. J Immunol. 2005. PMID: 16081841
-
Bcl11a is essential for lymphoid development and negatively regulates p53.J Exp Med. 2012 Dec 17;209(13):2467-83. doi: 10.1084/jem.20121846. Epub 2012 Dec 10. J Exp Med. 2012. PMID: 23230003 Free PMC article.
-
Ikaros fingers on lymphocyte differentiation.Int J Hematol. 2014 Sep;100(3):220-9. doi: 10.1007/s12185-014-1644-5. Epub 2014 Aug 2. Int J Hematol. 2014. PMID: 25085254 Free PMC article. Review.
-
Human innate lymphoid cells.Immunol Lett. 2016 Nov;179:2-8. doi: 10.1016/j.imlet.2016.01.007. Epub 2016 Feb 1. Immunol Lett. 2016. PMID: 26844414 Review.
Cited by
-
Cysteine facilitates the lignocellulolytic response of Trichoderma guizhouense NJAU4742 by indirectly up-regulating membrane sugar transporters.Biotechnol Biofuels Bioprod. 2023 Oct 27;16(1):159. doi: 10.1186/s13068-023-02418-9. Biotechnol Biofuels Bioprod. 2023. PMID: 37891614 Free PMC article.
-
Kitlo hematopoietic stem cells exhibit distinct lymphoid-primed chromatin landscapes that enhance thymic reconstitution.Nat Commun. 2025 Jul 4;16(1):6170. doi: 10.1038/s41467-025-61125-1. Nat Commun. 2025. PMID: 40615375 Free PMC article.
-
BTB protein family and human breast cancer: signaling pathways and clinical progress.J Cancer Res Clin Oncol. 2023 Nov;149(17):16213-16229. doi: 10.1007/s00432-023-05314-9. Epub 2023 Sep 8. J Cancer Res Clin Oncol. 2023. PMID: 37682360 Free PMC article. Review.
-
Effects of the Zbtb1 Gene on Chromatin Spatial Structure and Lymphatic Development: Combined Analysis of Hi-C, ATAC-Seq and RNA-Seq.Front Cell Dev Biol. 2022 Apr 25;10:874525. doi: 10.3389/fcell.2022.874525. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35547816 Free PMC article.
-
Rare variant analysis in eczema identifies exonic variants in DUSP1, NOTCH4 and SLC9A4.Nat Commun. 2021 Nov 16;12(1):6618. doi: 10.1038/s41467-021-26783-x. Nat Commun. 2021. PMID: 34785669 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

