Are Protein Force Fields Getting Better? A Systematic Benchmark on 524 Diverse NMR Measurements
- PMID: 22754404
- PMCID: PMC3383641
- DOI: 10.1021/ct2007814
Are Protein Force Fields Getting Better? A Systematic Benchmark on 524 Diverse NMR Measurements
Abstract
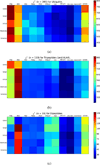
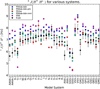
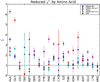
Recent hardware and software advances have enabled simulation studies of protein systems on biophysically-relevant timescales, often revealing the need for improved force fields. Although early force field development was limited by the lack of direct comparisons between simulation and experiment, recent work from several labs has demonstrated direct calculation of NMR observables from protein simulations. Here we quantitatively evaluate recent molecular dynamics force fields against a suite of 524 chemical shift and J coupling ((3)JH(N)H(α), (3)JH(N)C(β), (3)JH(α)C', (3)JH(N)C', and (3)JH(α)N) measurements on dipeptides, tripeptides, tetra-alanine, and ubiquitin. Of the force fields examined (ff96, ff99, ff03, ff03*, ff03w, ff99sb*, ff99sb-ildn, ff99sb-ildn-phi, ff99sb-ildn-nmr, CHARMM27, OPLS-AA), two force fields (ff99sb-ildn-phi, ff99sb-ildn-nmr) combining recent side chain and backbone torsion modifications achieve high accuracy in our benchmark. For the two optimal force fields, the calculation error is comparable to the uncertainty in the experimental comparison. This observation suggests that extracting additional force field improvements from NMR data may require increased accuracy in J coupling and chemical shift prediction. To further investigate the limitations of current force fields, we also consider conformational populations of dipeptides, which were recently estimated using vibrational spectroscopy.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
