An Integrated Bioinformatics and Computational Biology Approach Identifies New BH3-Only Protein Candidates
- PMID: 22754595
- PMCID: PMC3384560
- DOI: 10.2174/1874196701205010006
An Integrated Bioinformatics and Computational Biology Approach Identifies New BH3-Only Protein Candidates
Abstract
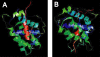
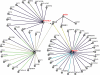
In this study, we utilized an integrated bioinformatics and computational biology approach in search of new BH3-only proteins belonging to the BCL2 family of apoptotic regulators. The BH3 (BCL2 homology 3) domain mediates specific binding interactions among various BCL2 family members. It is composed of an amphipathic α-helical region of approximately 13 residues that has only a few amino acids that are highly conserved across all members. Using a generalized motif, we performed a genome-wide search for novel BH3-containing proteins in the NCBI Consensus Coding Sequence (CCDS) database. In addition to known pro-apoptotic BH3-only proteins, 197 proteins were recovered that satisfied the search criteria. These were categorized according to α-helical content and predictive binding to BCL-xL (encoded by BCL2L1) and MCL-1, two representative anti-apoptotic BCL2 family members, using position-specific scoring matrix models. Notably, the list is enriched for proteins associated with autophagy as well as a broad spectrum of cellular stress responses such as endoplasmic reticulum stress, oxidative stress, antiviral defense, and the DNA damage response. Several potential novel BH3-containing proteins are highlighted. In particular, the analysis strongly suggests that the apoptosis inhibitor and DNA damage response regulator, AVEN, which was originally isolated as a BCL-xL-interacting protein, is a functional BH3-only protein representing a distinct subclass of BCL2 family members.
Figures


Similar articles
-
Binding affinity of pro-apoptotic BH3 peptides for the anti-apoptotic Mcl-1 and A1 proteins: Molecular dynamics simulations of Mcl-1 and A1 in complex with six different BH3 peptides.J Mol Graph Model. 2017 May;73:115-128. doi: 10.1016/j.jmgm.2016.12.006. Epub 2017 Feb 9. J Mol Graph Model. 2017. PMID: 28279820
-
Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.Autophagy. 2019 May;15(5):785-795. doi: 10.1080/15548627.2018.1564557. Epub 2019 Jan 9. Autophagy. 2019. PMID: 30626284 Free PMC article.
-
Computational Design of BH3-Mimetic Peptide Inhibitors That Can Bind Specifically to Mcl-1 or Bcl-XL: Role of Non-Hot Spot Residues.Biochemistry. 2020 Nov 17;59(45):4379-4394. doi: 10.1021/acs.biochem.0c00661. Epub 2020 Nov 4. Biochemistry. 2020. PMID: 33146015
-
AMBRA1, a Novel BH3-Like Protein: New Insights Into the AMBRA1-BCL2-Family Proteins Relationship.Int Rev Cell Mol Biol. 2017;330:85-113. doi: 10.1016/bs.ircmb.2016.09.002. Epub 2016 Nov 15. Int Rev Cell Mol Biol. 2017. PMID: 28215535 Review.
-
Structural biology of the Bcl-2 family of proteins.Biochim Biophys Acta. 2004 Mar 1;1644(2-3):83-94. doi: 10.1016/j.bbamcr.2003.08.012. Biochim Biophys Acta. 2004. PMID: 14996493 Review.
Cited by
-
Alterations of the interactome of Bcl-2 proteins in breast cancer at the transcriptional, mutational and structural level.PLoS Comput Biol. 2019 Dec 11;15(12):e1007485. doi: 10.1371/journal.pcbi.1007485. eCollection 2019 Dec. PLoS Comput Biol. 2019. PMID: 31825969 Free PMC article.
-
The novel BH3 α-helix mimetic JY-1-106 induces apoptosis in a subset of cancer cells (lung cancer, colon cancer and mesothelioma) by disrupting Bcl-xL and Mcl-1 protein-protein interactions with Bak.Mol Cancer. 2013 May 16;12(1):42. doi: 10.1186/1476-4598-12-42. Mol Cancer. 2013. PMID: 23680104 Free PMC article.
-
Redefining the BH3 Death Domain as a 'Short Linear Motif'.Trends Biochem Sci. 2015 Dec;40(12):736-748. doi: 10.1016/j.tibs.2015.09.007. Epub 2015 Nov 3. Trends Biochem Sci. 2015. PMID: 26541461 Free PMC article. Review.
-
Telomerase Reverse Transcriptase Contains a BH3-Like Motif and Interacts with BCL-2 Family Members.Mol Cells. 2018 Jul 31;41(7):684-694. doi: 10.14348/molcells.2018.0206. Epub 2018 Jun 25. Mol Cells. 2018. PMID: 29937479 Free PMC article.
-
Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes.Elife. 2015 Aug 12;4:e06234. doi: 10.7554/eLife.06234. Elife. 2015. PMID: 26267306 Free PMC article.
References
-
- Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW. BH3-only proteins: Orchestrators of apoptosis. Biochim Biophys Acta. 2011;1813:508–520. - PubMed
-
- Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. - PubMed
-
- Lama D, Sankararamakrishnan R. Molecular dynamics simulations of pro-apoptotic BH3 peptide helices in aqueous medium: relationship between helix stability and their binding affinities to the anti-apoptotic protein Bcl-X(L). J Comput Aided Mol Des. 2011;25:413–426. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
