Herpes virus fusion and entry: a story with many characters
- PMID: 22754650
- PMCID: PMC3386629
- DOI: 10.3390/v4050800
Herpes virus fusion and entry: a story with many characters
Abstract
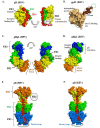
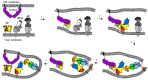
Herpesviridae comprise a large family of enveloped DNA viruses all of whom employ orthologs of the same three glycoproteins, gB, gH and gL. Additionally, herpesviruses often employ accessory proteins to bind receptors and/or bind the heterodimer gH/gL or even to determine cell tropism. Sorting out how these proteins function has been resolved to a large extent by structural biology coupled with supporting biochemical and biologic evidence. Together with the G protein of vesicular stomatitis virus, gB is a charter member of the Class III fusion proteins. Unlike VSV G, gB only functions when partnered with gH/gL. However, gH/gL does not resemble any known viral fusion protein and there is evidence that its function is to upregulate the fusogenic activity of gB. In the case of herpes simplex virus, gH/gL itself is upregulated into an active state by the conformational change that occurs when gD, the receptor binding protein, binds one of its receptors. In this review we focus primarily on prototypes of the three subfamilies of herpesviruses. We will present our model for how herpes simplex virus (HSV) regulates fusion in series of highly regulated steps. Our model highlights what is known and also provides a framework to address mechanistic questions about fusion by HSV and herpesviruses in general.
Keywords: CMV; EBV; HSV; VZV; crystal structure; functional region; glycoproteins; monoclonal antibody.
Figures



References
-
- Campadelli-Fiume G., Menotti L. Entry of Alphaherpesviruses into the Cell. In: Arvin A., Campadelli-Fiume G., Mocarski E., Moore P.S., Roizman B., Whitley R., Yamanishi K., editors. Source Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge University Press; Cambridge, UK: 2007. Chapter 7. - PubMed
-
- Campadelli-Fiume G., Menotti L., Avitabile E., Gianni T. Viral and cellular contributions to herpes simplex virus entry into the cell. Curr. Opin. Virol. 2012;2:28–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

