Latest insights on adenovirus structure and assembly
- PMID: 22754652
- PMCID: PMC3386624
- DOI: 10.3390/v4050847
Latest insights on adenovirus structure and assembly
Erratum in
- Viruses. 2012;4(12):3952
Abstract
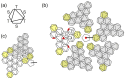
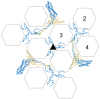
Adenovirus (AdV) capsid organization is considerably complex, not only because of its large size (~950 Å) and triangulation number (pseudo T = 25), but also because it contains four types of minor proteins in specialized locations modulating the quasi-equivalent icosahedral interactions. Up until 2009, only its major components (hexon, penton, and fiber) had separately been described in atomic detail. Their relationships within the virion, and the location of minor coat proteins, were inferred from combining the known crystal structures with increasingly more detailed cryo-electron microscopy (cryoEM) maps. There was no structural information on assembly intermediates. Later on that year, two reports described the structural differences between the mature and immature adenoviral particle, starting to shed light on the different stages of viral assembly, and giving further insights into the roles of core and minor coat proteins during morphogenesis [1,2]. Finally, in 2010, two papers describing the atomic resolution structure of the complete virion appeared [3,4]. These reports represent a veritable tour de force for two structural biology techniques: X-ray crystallography and cryoEM, as this is the largest macromolecular complex solved at high resolution by either of them. In particular, the cryoEM analysis provided an unprecedented clear picture of the complex protein networks shaping the icosahedral shell. Here I review these latest developments in the field of AdV structural studies.
Keywords: adenovirus; core proteins; cryo-electron microscopy; crystallography; maturation; minor coat proteins; structure.
Figures






References
-
- Berk A.J. Adenoviridae: The Viruses and Their Replication. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., editors. Fields Virology. 5th. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 2355–2394.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

