Programmed cell death 4 (PDCD4): a novel player in ethanol-mediated suppression of protein translation in primary cortical neurons and developing cerebral cortex
- PMID: 22757755
- PMCID: PMC3724449
- DOI: 10.1111/j.1530-0277.2012.01850.x
Programmed cell death 4 (PDCD4): a novel player in ethanol-mediated suppression of protein translation in primary cortical neurons and developing cerebral cortex
Abstract
Background: Prenatal exposure to ethanol (EtOH) elicits a range of neuro-developmental abnormalities, microcephaly to behavioral deficits. Impaired protein synthesis has been connected to pathogenesis of EtOH-induced brain damage and abnormal neuron development. However, mechanisms underlying these impairments of protein synthesis are not known. In this study, we illustrate the effects of EtOH on programmed cell death protein 4 (PDCD4), a tumor and translation repressor.
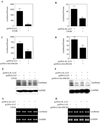
Methods: Primary cortical neurons (PCNs) were treated with 2.5 and 4 mg/ml EtOH for different time points (4 to 24 hours), and PDCD4 expression was detected by Western blotting. Protein synthesis was determined using [(35) S] methionine incorporation assay. Methyl cap pull-down assay was performed to establish the effect of EtOH on association of eukaryotic initiation factor 4A (eIF4A) with capped mRNA. Luciferase assay was performed to determine the in vivo translation. A 2-day acute 5-dose binge model with EtOH (4 g/kg body wt, 25% v/v) was performed in Sprague-Dawley rats at 12-hour intervals and analyzed for PDCD4, eIF4A, and eIF4A-methyl cap association.
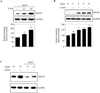
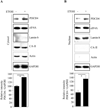
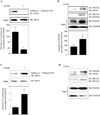
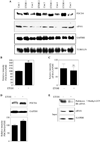
Results: EtOH increased PDCD4 expression in a time- and dose-dependent manner in PCNs, which inhibited the association of eIF4A with methyl cap. EtOH and ectopic PDCD4 expression suppressed in vivo translation in PCNs and RNAi targeting of PDCD4 blocked the inhibitory effect of EtOH on protein synthesis. In utero exposure of pregnant rats to EtOH resulted in a significant increase in PDCD4 in fetal cerebral cortex along with the inhibition of methyl cap-associated eIF4A, compared with isocaloric controls. Increased PDCD4 also occurred in pooled fractions of remaining brain regions.
Conclusions: Our data, for the first time, illustrate that PDCD4 mediates inhibitory effects of EtOH on protein synthesis in PCNs and developing brain.
Copyright © 2012 by the Research Society on Alcoholism.
Figures

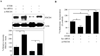

Similar articles
-
Programmed cell death 4 mechanism of action: The model to be updated?Cell Cycle. 2017 Oct 2;16(19):1761-1764. doi: 10.1080/15384101.2017.1371881. Epub 2017 Aug 30. Cell Cycle. 2017. PMID: 28853972 Free PMC article.
-
Ethanol-induced transcriptional activation of programmed cell death 4 (Pdcd4) is mediated by GSK-3β signaling in rat cortical neuroblasts.PLoS One. 2014 May 16;9(5):e98080. doi: 10.1371/journal.pone.0098080. eCollection 2014. PLoS One. 2014. PMID: 24837604 Free PMC article.
-
The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation.Mol Cell Biol. 2003 Jan;23(1):26-37. doi: 10.1128/MCB.23.1.26-37.2003. Mol Cell Biol. 2003. PMID: 12482958 Free PMC article.
-
Ethanol metabolism and effects: nitric oxide and its interaction.Curr Clin Pharmacol. 2007 May;2(2):145-53. doi: 10.2174/157488407780598135. Curr Clin Pharmacol. 2007. PMID: 18690862 Free PMC article. Review.
-
The Impact of Pdcd4, a Translation Inhibitor, on Drug Resistance.Pharmaceuticals (Basel). 2024 Oct 19;17(10):1396. doi: 10.3390/ph17101396. Pharmaceuticals (Basel). 2024. PMID: 39459035 Free PMC article. Review.
Cited by
-
PDCD4 regulates axonal growth by translational repression of neurite growth-related genes and is modulated during nerve injury responses.RNA. 2020 Nov;26(11):1637-1653. doi: 10.1261/rna.075424.120. Epub 2020 Aug 3. RNA. 2020. PMID: 32747606 Free PMC article.
-
Programmed cell death 4 as an endogenous suppressor of BDNF translation is involved in stress-induced depression.Mol Psychiatry. 2021 Jun;26(6):2316-2333. doi: 10.1038/s41380-020-0692-x. Epub 2020 Mar 16. Mol Psychiatry. 2021. PMID: 32203159 Free PMC article.
-
Neuronal activity regulates the nuclear proteome to promote activity-dependent transcription.J Cell Biol. 2021 Dec 6;220(12):e202103087. doi: 10.1083/jcb.202103087. Epub 2021 Oct 7. J Cell Biol. 2021. PMID: 34617965 Free PMC article.
-
Ethanol (E) Impairs Fetal Brain GSH Homeostasis by Inhibiting Excitatory Amino-Acid Carrier 1 (EAAC1)-Mediated Cysteine Transport.Int J Mol Sci. 2017 Dec 5;18(12):2596. doi: 10.3390/ijms18122596. Int J Mol Sci. 2017. PMID: 29206135 Free PMC article.
-
Translating from cancer to the brain: regulation of protein synthesis by eIF4F.Learn Mem. 2019 Aug 15;26(9):332-342. doi: 10.1101/lm.050047.119. Print 2019 Sep. Learn Mem. 2019. PMID: 31416906 Free PMC article. Review.
References
-
- Archibald SL, Fennema-Notestine C, Gamst A, Riley EP, Mattson SN, Jernigan TL. Brain dysmorphology in individuals with severe prenatal alcohol exposure. Dev Med Child Neurol. 2001;43:148–154. - PubMed
-
- Böhm M, Sawicka K, Siebrasse JP, Brehmer-Fastnacht A, Peters R, Klempnauer KH. The transformation suppressor protein Pdcd4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene. 2003;22:4905–4910. - PubMed
-
- Bonner AB, Dalwai S, Marway JS, Preedy VR. Acute exposure to the nutritional toxin alcohol reduces brain protein synthesis in vivo. Metabolism. 2003;52:389–396. - PubMed
-
- Bookstein FL, Streissguth AP, Sampson PD, Connor PD, Barr HM. Corpus callosum shape and neuropsychological deficits in adult males with heavy fetal alcohol exposure. Neuroimage. 2002;15:233–251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

