Catalytic and structural role of a conserved active site histidine in berberine bridge enzyme
- PMID: 22757961
- PMCID: PMC3413249
- DOI: 10.1021/bi300411n
Catalytic and structural role of a conserved active site histidine in berberine bridge enzyme
Abstract
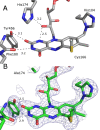
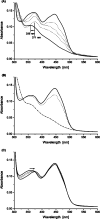
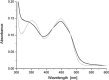
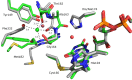
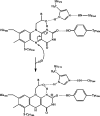
Berberine bridge enzyme (BBE) is a paradigm for the class of bicovalently flavinylated oxidases, which catalyzes the oxidative cyclization of (S)-reticuline to (S)-scoulerine. His174 was identified as an important active site residue because of its role in the stabilization of the reduced state of the flavin cofactor. It is also strictly conserved in the family of BBE-like oxidases. Here, we present a detailed biochemical and structural characterization of a His174Ala variant supporting its importance during catalysis and for the structural organization of the active site. Substantial changes in all kinetic parameters and a decrease in midpoint potential were observed for the BBE His174Ala variant protein. Moreover, the crystal structure of the BBE His174Ala variant showed significant structural rearrangements compared to wild-type enzyme. On the basis of our findings, we propose that His174 is part of a hydrogen bonding network that stabilizes the negative charge at the N1-C2=O locus via interaction with the hydroxyl group at C2' of the ribityl side chain of the flavin cofactor. Hence, replacement of this residue with alanine reduces the stabilizing effect for the transiently formed negative charge and results in drastically decreased kinetic parameters as well as a lower midpoint redox potential.
Figures
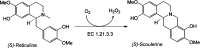


Similar articles
-
Structural and mechanistic studies reveal the functional role of bicovalent flavinylation in berberine bridge enzyme.J Biol Chem. 2009 Jul 24;284(30):19993-20001. doi: 10.1074/jbc.M109.015727. Epub 2009 May 19. J Biol Chem. 2009. PMID: 19457868 Free PMC article.
-
A concerted mechanism for berberine bridge enzyme.Nat Chem Biol. 2008 Dec;4(12):739-41. doi: 10.1038/nchembio.123. Epub 2008 Oct 26. Nat Chem Biol. 2008. PMID: 18953357
-
The catalytic machinery of the FAD-dependent AtBBE-like protein 15 for alcohol oxidation: Y193 and Y479 form a catalytic base, Q438 and R292 an alkoxide binding site.Arch Biochem Biophys. 2021 Mar 30;700:108766. doi: 10.1016/j.abb.2021.108766. Epub 2021 Jan 22. Arch Biochem Biophys. 2021. PMID: 33485849
-
The family of berberine bridge enzyme-like enzymes: A treasure-trove of oxidative reactions.Arch Biochem Biophys. 2017 Oct 15;632:88-103. doi: 10.1016/j.abb.2017.06.023. Epub 2017 Jul 1. Arch Biochem Biophys. 2017. PMID: 28676375 Review.
-
Biosynthetic Strategies of Berberine Bridge Enzyme-like Flavoprotein Oxidases toward Structural Diversification in Natural Product Biosynthesis.Biochemistry. 2024 Sep 3;63(17):2089-2110. doi: 10.1021/acs.biochem.4c00320. Epub 2024 Aug 12. Biochemistry. 2024. PMID: 39133819 Free PMC article. Review.
Cited by
-
Plant-like biosynthesis of isoquinoline alkaloids in Aspergillus fumigatus.Nat Chem Biol. 2016 Jun;12(6):419-24. doi: 10.1038/nchembio.2061. Epub 2016 Apr 11. Nat Chem Biol. 2016. PMID: 27065235 Free PMC article.
-
The 2'-hydroxy group of flavin mononucleotide influences the catalytic function and promiscuity of the flavoprotein iodotyrosine dehalogenase.RSC Chem Biol. 2023 Aug 4;4(9):698-705. doi: 10.1039/d3cb00094j. eCollection 2023 Aug 30. RSC Chem Biol. 2023. PMID: 37654510 Free PMC article.
-
Advances in the biosynthesis of naturally occurring benzylisoquinoline alkaloids.Front Plant Sci. 2025 Jan 30;16:1548471. doi: 10.3389/fpls.2025.1548471. eCollection 2025. Front Plant Sci. 2025. PMID: 39949415 Free PMC article. Review.
-
The Flavoproteome of the Model Plant Arabidopsis thaliana.Int J Mol Sci. 2020 Jul 28;21(15):5371. doi: 10.3390/ijms21155371. Int J Mol Sci. 2020. PMID: 32731628 Free PMC article.
-
Oxidative Cyclization in Natural Product Biosynthesis.Chem Rev. 2017 Apr 26;117(8):5226-5333. doi: 10.1021/acs.chemrev.6b00478. Epub 2016 Dec 12. Chem Rev. 2017. PMID: 27936626 Free PMC article. Review.
References
-
- Winkler A.; Hartner F.; Kutchan T. M.; Glieder A.; Macheroux P. (2006) Biochemical evidence that berberine bridge enzyme belongs to a novel family of flavoproteins containing a bi-covalently attached FAD cofactor. J. Biol. Chem. 281, 21276–21285. - PubMed
-
- Heuts D. P. H. M.; Winter R. T.; Damsma G. E.; Janssen D. B.; Fraaije M. W. (2008) The role of double covalent flavin binding in chito-oligosaccharide oxidase from Fusarium graminearum. Biochem. J. 413, 175–183. - PubMed
-
- Mo X.; Huang H.; Ma J.; Wang Z.; Wang B.; Zhang S.; Zhang C.; Ju J. (2011) Characterization of TrdL as a 10-hydroxy dehydrogenase and generation of new analogues from a tirandamycin biosynthetic pathway. Org. Lett. 13, 2212–2215. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

