BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence
- PMID: 22757963
- PMCID: PMC3402693
- DOI: 10.1111/j.1365-2958.2012.08127.x
BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence
Abstract
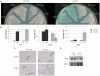
In the opportunistic fungal pathogen Candida albicans both cellular morphology and the capacity to cause disease are regulated by the transcriptional repressor Nrg1p. One of the genes repressed by Nrg1p is BRG1, which encodes a putative GATA family transcription factor. Deletion of both copies of this gene prevents hypha formation. We discovered that BRG1 overexpression is sufficient to overcome Nrg1p-mediated repression and drive the morphogenetic shift from yeast to hyphae even in the absence of environmental stimuli. We further observed that expression of BRG1 influences the stability of the NRG1 transcript, thus controlling filamentation through a feedback loop. Analysis of this phenomenon revealed that BRG1 expression is required for the induction of an antisense NRG1 transcript. This is the first demonstration of a role for mRNA stability in regulating the key C. albicans virulence trait: the ability to form hyphae.
© 2012 Blackwell Publishing Ltd.
Figures




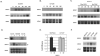


References
-
- Bendel CM, Hess DJ, Garni RM, Henry-Stanley M, Wells CL. Comparative virulence of Candida albicans yeast and filamentous forms in orally and intravenously inoculated mice. Crit Care Med. 2003;31:501–507. - PubMed
-
- Boland JM, Chung HH, Robberts FJ, Wilson WR, Steckelberg JM, Baddour LM, Miller DV. Fungal prosthetic valve endocarditis: Mayo Clinic experience with a clinicopathological analysis. Mycoses. 2010 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

