An inventory of peroxisomal proteins and pathways in Drosophila melanogaster
- PMID: 22758915
- PMCID: PMC3443258
- DOI: 10.1111/j.1600-0854.2012.01393.x
An inventory of peroxisomal proteins and pathways in Drosophila melanogaster
Abstract
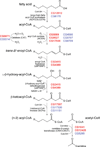
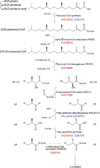
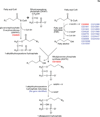
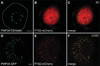
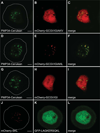
Peroxisomes are ubiquitous organelles housing a variety of essential biochemical pathways. Peroxisome dysfunction causes a spectrum of human diseases known as peroxisome biogenesis disorders (PBD). Although much is known regarding the mechanism of peroxisome biogenesis, it is still unclear how peroxisome dysfunction leads to the disease state. Several recent studies have shown that mutations in Drosophila peroxin genes cause phenotypes similar to those seen in humans with PBDs suggesting that Drosophila might be a useful system to model PBDs. We have analyzed the proteome of Drosophila to identify the proteins involved in peroxisomal biogenesis and homeostasis as well as metabolic enzymes that function within the organelle. The subcellular localization of five of these predicted peroxisomal proteins was confirmed. Similar to Caenorhabditis elegans, Drosophila appears to only utilize the peroxisome targeting signal type 1 system for matrix protein import. This work will further our understanding of peroxisomes in Drosophila and add to the usefulness of this emerging model system.
© 2012 John Wiley & Sons A/S.
Figures







Similar articles
-
A Systematic Cell-Based Analysis of Localization of Predicted Drosophila Peroxisomal Proteins.Traffic. 2016 May;17(5):536-53. doi: 10.1111/tra.12384. Epub 2016 Mar 1. Traffic. 2016. PMID: 26865094
-
Peroxisomes are required for lipid metabolism and muscle function in Drosophila melanogaster.PLoS One. 2014 Jun 19;9(6):e100213. doi: 10.1371/journal.pone.0100213. eCollection 2014. PLoS One. 2014. PMID: 24945818 Free PMC article.
-
Peroxisome Protein Prediction in Drosophila melanogaster.Subcell Biochem. 2018;89:235-258. doi: 10.1007/978-981-13-2233-4_10. Subcell Biochem. 2018. PMID: 30378026 Review.
-
Molecular basis of peroxisomal biogenesis disorders caused by defects in peroxisomal matrix protein import.Biochim Biophys Acta. 2012 Sep;1822(9):1326-36. doi: 10.1016/j.bbadis.2012.05.010. Epub 2012 May 19. Biochim Biophys Acta. 2012. PMID: 22617146 Review.
-
A Drosophila model for the Zellweger spectrum of peroxisome biogenesis disorders.Dis Model Mech. 2011 Sep;4(5):659-72. doi: 10.1242/dmm.007419. Epub 2011 Jun 13. Dis Model Mech. 2011. PMID: 21669930 Free PMC article.
Cited by
-
Rosy Beginnings: Studying Peroxisomes in Drosophila.Front Cell Dev Biol. 2020 Aug 25;8:835. doi: 10.3389/fcell.2020.00835. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32984330 Free PMC article. Review.
-
Translating the Arabidopsis thaliana Peroxisome Proteome Insights to Solanum lycopersicum: Consensus Versus Diversity.Front Cell Dev Biol. 2022 Jul 13;10:909604. doi: 10.3389/fcell.2022.909604. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35912119 Free PMC article. Review.
-
Adaptation to Chronic Nutritional Stress Leads to Reduced Dependence on Microbiota in Drosophila melanogaster.mBio. 2017 Oct 24;8(5):e01496-17. doi: 10.1128/mBio.01496-17. mBio. 2017. PMID: 29066546 Free PMC article.
-
The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance.Front Physiol. 2015 Sep 24;6:259. doi: 10.3389/fphys.2015.00259. eCollection 2015. Front Physiol. 2015. PMID: 26441678 Free PMC article. Review.
-
Unbalanced lipolysis results in lipotoxicity and mitochondrial damage in peroxisome-deficient Pex19 mutants.Mol Biol Cell. 2018 Feb 15;29(4):396-407. doi: 10.1091/mbc.E17-08-0535. Epub 2017 Dec 27. Mol Biol Cell. 2018. PMID: 29282281 Free PMC article.
References
-
- Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. - PubMed
-
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122(1):85–95. - PubMed
-
- Platta HW, Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17(10):474–484. - PubMed
-
- Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105(2):187–196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

