M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins
- PMID: 22759232
- PMCID: PMC3564602
- DOI: 10.1021/bc300130z
M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins
Abstract
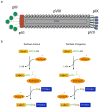
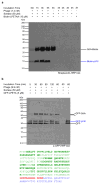
We exploit bacterial sortases to attach a variety of moieties to the capsid proteins of M13 bacteriophage. We show that pIII, pIX, and pVIII can be functionalized with entities ranging from small molecules (e.g., fluorophores, biotin) to correctly folded proteins (e.g., GFP, antibodies, streptavidin) in a site-specific manner, and with yields that surpass those of any reported using phage display technology. A case in point is modification of pVIII. While a phage vector limits the size of the insert into pVIII to a few amino acids, a phagemid system limits the number of copies actually displayed at the surface of M13. Using sortase-based reactions, a 100-fold increase in the efficiency of display of GFP onto pVIII is achieved. Taking advantage of orthogonal sortases, we can simultaneously target two distinct capsid proteins in the same phage particle and maintain excellent specificity of labeling. As demonstrated in this work, this is a simple and effective method for creating a variety of structures, thus expanding the use of M13 for materials science applications and as a biological tool.
Figures




References
-
- Sidhu SS. Engineering M13 for phage display. Biomol Eng. 2001;18:57–63. - PubMed
-
- Burritt JB, Quinn MT, Jutila MA, Bond CW, Jesaitis AJ. Topological mapping of neutrophil cytochrome b epitopes with phage-display libraries. J Biol Chem. 1995;270:16974–80. - PubMed
-
- Barry MA, Dower WJ, Johnston SA. Toward cell-targeting gene therapy vectors: selection of cell-binding peptides from random peptide-presenting phage libraries. Nat Med. 1996;2:299–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

