The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage
- PMID: 22759610
- PMCID: PMC3394424
- DOI: 10.1186/1471-2164-13-S3-S1
The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage
Abstract
Background: Phenylalanine ammonia lyase (PAL) is a key enzyme of the phenylpropanoid pathway that catalyzes the deamination of phenylalanine to trans-cinnamic acid, a precursor for the lignin and flavonoid biosynthetic pathways. To date, PAL genes have been less extensively studied in gymnosperms than in angiosperms. Our interest in PAL genes stems from their potential role in the defense responses of Pinus taeda, especially with respect to lignification and production of low molecular weight phenolic compounds under various biotic and abiotic stimuli. In contrast to all angiosperms for which reference genome sequences are available, P. taeda has previously been characterized as having only a single PAL gene. Our objective was to re-evaluate this finding, assess the evolutionary history of PAL genes across major angiosperm and gymnosperm lineages, and characterize PAL gene expression patterns in Pinus taeda.
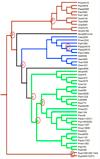
Methods: We compiled a large set of PAL genes from the largest transcript dataset available for P. taeda and other conifers. The transcript assemblies for P. taeda were validated through sequencing of PCR products amplified using gene-specific primers based on the putative PAL gene assemblies. Verified PAL gene sequences were aligned and a gene tree was estimated. The resulting gene tree was reconciled with a known species tree and the time points for gene duplication events were inferred relative to the divergence of major plant lineages.
Results: In contrast to angiosperms, gymnosperms have retained a diverse set of PAL genes distributed among three major clades that arose from gene duplication events predating the divergence of these two seed plant lineages. Whereas multiple PAL genes have been identified in sequenced angiosperm genomes, all characterized angiosperm PAL genes form a single clade in the gene PAL tree, suggesting they are derived from a single gene in an ancestral angiosperm genome. The five distinct PAL genes detected and verified in P. taeda were derived from a combination of duplication events predating and postdating the divergence of angiosperms and gymnosperms.
Conclusions: Gymnosperms have a more phylogenetically diverse set of PAL genes than angiosperms. This inference has contrasting implications for the evolution of PAL gene function in gymnosperms and angiosperms.
Figures

References
-
- de Laubenfels DJ. The status of "conifers" in vegetation classifications. Ann Assoc Am Geogr. 1957;47:145–149. doi: 10.1111/j.1467-8306.1957.tb01530.x. - DOI
-
- Bonello P, Gordon TR, Herms DA, Wood DL, Erbilgin N. Nature and ecological implications of pathogen-induced systemic resistance in conifers: a novel hypothesis. Physiol Mol Plant Pathol. 2006;68:95–104. doi: 10.1016/j.pmpp.2006.12.002. - DOI
-
- Adomas A, Heller G, Li G, Olson A, Chu T, Osborne J, Craig D, Zyl LV, Wolfinger R, Sederoff R, Dean RA, Stenlid J, Finlay R, Asiegbu FO. Transcript profiling of a conifer pathosystem: response of Pinus sylvestris root tissues to pathogen (Heterobasidion annosum) invasion. Tree Physiol. 2007;27:1441–1458. doi: 10.1093/treephys/27.10.1441. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

