Multi-objective dynamic population shuffled frog-leaping biclustering of microarray data
- PMID: 22759615
- PMCID: PMC3394423
- DOI: 10.1186/1471-2164-13-S3-S6
Multi-objective dynamic population shuffled frog-leaping biclustering of microarray data
Abstract
Background: Multi-objective optimization (MOO) involves optimization problems with multiple objectives. Generally, theose objectives is used to estimate very different aspects of the solutions, and these aspects are often in conflict with each other. MOO first gets a Pareto set, and then looks for both commonality and systematic variations across the set. For the large-scale data sets, heuristic search algorithms such as EA combined with MOO techniques are ideal. Newly DNA microarray technology may study the transcriptional response of a complete genome to different experimental conditions and yield a lot of large-scale datasets. Biclustering technique can simultaneously cluster rows and columns of a dataset, and hlep to extract more accurate information from those datasets. Biclustering need optimize several conflicting objectives, and can be solved with MOO methods. As a heuristics-based optimization approach, the particle swarm optimization (PSO) simulate the movements of a bird flock finding food. The shuffled frog-leaping algorithm (SFL) is a population-based cooperative search metaphor combining the benefits of the local search of PSO and the global shuffled of information of the complex evolution technique. SFL is used to solve the optimization problems of the large-scale datasets.
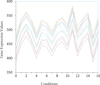
Results: This paper integrates dynamic population strategy and shuffled frog-leaping algorithm into biclustering of microarray data, and proposes a novel multi-objective dynamic population shuffled frog-leaping biclustering (MODPSFLB) algorithm to mine maximum bicluesters from microarray data. Experimental results show that the proposed MODPSFLB algorithm can effectively find significant biological structures in terms of related biological processes, components and molecular functions.
Conclusions: The proposed MODPSFLB algorithm has good diversity and fast convergence of Pareto solutions and will become a powerful systematic functional analysis in genome research.
Figures
References
-
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96(6):2907–2912. doi: 10.1073/pnas.96.6.2907. - DOI - PMC - PubMed
-
- Cheng Y, Church GM. Biclustering of expression data. Proc Int Conf Intell Syst Mol Biol. 2000;8(1):93–103. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

