ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response
- PMID: 22759634
- PMCID: PMC3404384
- DOI: 10.1101/gad.193516.112
ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response
Abstract
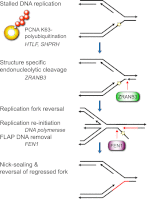
To efficiently duplicate their genomic content, cells must overcome DNA lesions that interfere with processive DNA replication. These lesions may be removed and repaired, rather than just tolerated, to allow continuity of DNA replication on an undamaged DNA template. However, it is unclear how this is achieved at a molecular level. Here we identify a new replication-associated factor, ZRANB3 (zinc finger, RAN-binding domain containing 3), and propose its role in the repair of replication-blocking lesions. ZRANB3 has a unique structure-specific endonuclease activity, which is coupled to ATP hydrolysis. It cleaves branched DNA structures with unusual polarity, generating an accessible 3'-OH group in the template of the leading strand. Furthermore, ZRANB3 localizes to DNA replication sites and interacts with the components of the replication machinery. It is recruited to damaged replication forks via multiple mechanisms, which involve interactions with PCNA, K63-polyubiquitin chains, and branched DNA structures. Collectively, our data support a role for ZRANB3 in the replication stress response and suggest new insights into how DNA repair is coordinated with DNA replication to maintain genome stability.
Figures



Comment in
-
Finally, polyubiquitinated PCNA gets recognized.Mol Cell. 2012 Aug 10;47(3):333-4. doi: 10.1016/j.molcel.2012.07.024. Mol Cell. 2012. PMID: 22883622
Similar articles
-
Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress.Mol Cell. 2012 Aug 10;47(3):396-409. doi: 10.1016/j.molcel.2012.05.024. Epub 2012 Jun 14. Mol Cell. 2012. PMID: 22704558 Free PMC article.
-
Structural insights into the function of ZRANB3 in replication stress response.Nat Commun. 2017 Jun 16;8:15847. doi: 10.1038/ncomms15847. Nat Commun. 2017. PMID: 28621305 Free PMC article.
-
Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity.Mol Cell. 2017 Sep 7;67(5):882-890.e5. doi: 10.1016/j.molcel.2017.08.010. Mol Cell. 2017. PMID: 28886337 Free PMC article.
-
Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability.Crit Rev Biochem Mol Biol. 2017 Dec;52(6):696-714. doi: 10.1080/10409238.2017.1380597. Epub 2017 Sep 28. Crit Rev Biochem Mol Biol. 2017. PMID: 28954549 Free PMC article. Review.
-
Nucleases Acting at Stalled Forks: How to Reboot the Replication Program with a Few Shortcuts.Annu Rev Genet. 2017 Nov 27;51:477-499. doi: 10.1146/annurev-genet-120116-024745. Annu Rev Genet. 2017. PMID: 29178820 Review.
Cited by
-
Homologous recombination maintenance of genome integrity during DNA damage tolerance.Mol Cell Oncol. 2014 Oct 29;1(2):e957039. doi: 10.4161/23723548.2014.957039. eCollection 2014 Apr-Jun. Mol Cell Oncol. 2014. PMID: 27308329 Free PMC article. Review.
-
PARP1 recruits DNA translocases to restrain DNA replication and facilitate DNA repair.PLoS Genet. 2022 Dec 13;18(12):e1010545. doi: 10.1371/journal.pgen.1010545. eCollection 2022 Dec. PLoS Genet. 2022. PMID: 36512630 Free PMC article.
-
Homologous recombination defects and how they affect replication fork maintenance.AIMS Genet. 2019 Apr 3;5(4):192-211. doi: 10.3934/genet.2018.4.192. eCollection 2018. AIMS Genet. 2019. PMID: 31435521 Free PMC article. Review.
-
Systematic classification of the His-Me finger superfamily.Nucleic Acids Res. 2017 Nov 16;45(20):11479-11494. doi: 10.1093/nar/gkx924. Nucleic Acids Res. 2017. PMID: 29040665 Free PMC article.
-
Maneuvers on PCNA Rings during DNA Replication and Repair.Genes (Basel). 2018 Aug 17;9(8):416. doi: 10.3390/genes9080416. Genes (Basel). 2018. PMID: 30126151 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
