Domain landscapes of somatic mutations in cancer
- PMID: 22759657
- PMCID: PMC3394412
- DOI: 10.1186/1471-2164-13-S4-S9
Domain landscapes of somatic mutations in cancer
Abstract
Background: Large-scale tumor sequencing projects are now underway to identify genetic mutations that drive tumor initiation and development. Most studies take a gene-based approach to identifying driver mutations, highlighting genes mutated in a large percentage of tumor samples as those likely to contain driver mutations. However, this gene-based approach usually does not consider the position of the mutation within the gene or the functional context the position of the mutation provides. Here we introduce a novel method for mapping mutations to distinct protein domains, not just individual genes, in which they occur, thus providing the functional context for how the mutation contributes to disease. Furthermore, aggregating mutations from all genes containing a specific protein domain enables the identification of mutations that are rare at the gene level, but that occur frequently within the specified domain. These highly mutated domains potentially reveal disruptions of protein function necessary for cancer development.
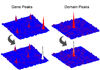
Results: We mapped somatic mutations from the protein coding regions of 100 colon adenocarcinoma tumor samples to the genes and protein domains in which they occurred, and constructed topographical maps to depict the "mutational landscapes" of gene and domain mutation frequencies. We found significant mutation frequency in a number of genes previously known to be somatically mutated in colon cancer patients including APC, TP53 and KRAS. In addition, we found significant mutation frequency within specific domains located in these genes, as well as within other domains contained in genes having low mutation frequencies. These domain "peaks" were enriched with functions important to cancer development including kinase activity, DNA binding and repair, and signal transduction.
Conclusions: Using our method to create the domain landscapes of mutations in colon cancer, we were able to identify somatic mutations with high potential to drive cancer development. Interestingly, the majority of the genes involved have a low mutation frequency. Therefore, the method shows good potential for identifying rare driver mutations in current, large-scale tumor sequencing projects. In addition, mapping mutations to specific domains provides the necessary functional context for understanding how the mutations contribute to the disease, and may reveal novel or more refined gene and domain target regions for drug development.
Figures




Similar articles
-
Frequency of Somatic TP53 Mutations in Combination with Known Pathogenic Mutations in Colon Adenocarcinoma, Non-Small Cell Lung Carcinoma, and Gliomas as Identified by Next-Generation Sequencing.Neoplasia. 2018 Mar;20(3):256-262. doi: 10.1016/j.neo.2017.12.005. Epub 2018 Feb 16. Neoplasia. 2018. PMID: 29454261 Free PMC article.
-
De novo discovery of mutated driver pathways in cancer.Genome Res. 2012 Feb;22(2):375-85. doi: 10.1101/gr.120477.111. Epub 2011 Jun 7. Genome Res. 2012. PMID: 21653252 Free PMC article.
-
Identification and analysis of mutational hotspots in oncogenes and tumour suppressors.Oncotarget. 2017 Mar 28;8(13):21290-21304. doi: 10.18632/oncotarget.15514. Oncotarget. 2017. PMID: 28423505 Free PMC article.
-
Whole-exome sequencing reveals recurrent somatic mutation networks in cancer.Cancer Lett. 2013 Nov 1;340(2):270-6. doi: 10.1016/j.canlet.2012.11.002. Epub 2012 Nov 12. Cancer Lett. 2013. PMID: 23153794 Review.
-
Combinatorial patterns of somatic gene mutations in cancer.FASEB J. 2008 Aug;22(8):2605-22. doi: 10.1096/fj.08-108985. Epub 2008 Apr 23. FASEB J. 2008. PMID: 18434431 Review.
Cited by
-
Pathway-specific protein domains are predictive for human diseases.PLoS Comput Biol. 2019 May 10;15(5):e1007052. doi: 10.1371/journal.pcbi.1007052. eCollection 2019 May. PLoS Comput Biol. 2019. PMID: 31075101 Free PMC article.
-
Analysis of individual protein regions provides novel insights on cancer pharmacogenomics.PLoS Comput Biol. 2015 Jan 8;11(1):e1004024. doi: 10.1371/journal.pcbi.1004024. eCollection 2015 Jan. PLoS Comput Biol. 2015. PMID: 25568936 Free PMC article.
-
SNP-SIG Meeting 2011: identification and annotation of SNPs in the context of structure, function, and disease.BMC Genomics. 2012 Jun 18;13 Suppl 4(Suppl 4):S1. doi: 10.1186/1471-2164-13-S4-S1. BMC Genomics. 2012. PMID: 22759647 Free PMC article. No abstract available.
-
Computational algorithms for in silico profiling of activating mutations in cancer.Cell Mol Life Sci. 2019 Jul;76(14):2663-2679. doi: 10.1007/s00018-019-03097-2. Epub 2019 Apr 13. Cell Mol Life Sci. 2019. PMID: 30982079 Free PMC article. Review.
-
Large-scale pathogenicity prediction analysis of cancer-associated kinase mutations reveals variability in sensitivity and specificity of computational methods.Cancer Med. 2023 Aug;12(16):17468-17474. doi: 10.1002/cam4.6324. Epub 2023 Jul 6. Cancer Med. 2023. PMID: 37409618 Free PMC article.
References
-
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Science. 5797. Vol. 314. New York, NY; 2006. The consensus coding sequences of human breast and colorectal cancers; pp. 268–274. - PubMed
-
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Science. 5853. Vol. 318. New York, NY; 2007. The genomic landscapes of human breast and colorectal cancers; pp. 1108–1113. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

