p23 co-chaperone protects the aryl hydrocarbon receptor from degradation in mouse and human cell lines
- PMID: 22759865
- PMCID: PMC3418383
- DOI: 10.1016/j.bcp.2012.06.018
p23 co-chaperone protects the aryl hydrocarbon receptor from degradation in mouse and human cell lines
Abstract
The aryl hydrocarbon receptor (AhR) is a ligand-sensitive transcription factor which is responsible for most 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicities. Without ligand, the AhR complex is cytoplasmic and contains p23. Our objective was to investigate whether the wild type p23 levels are important for the AhR function. We generated eight p23-specific knockdown stable cell lines via either electroporation or lentiviral infection. Five of these stable cell lines were generated from a mouse hepatoma cell line (Hepa1c1c7) and three were from human hepatoma and cervical cell lines (Hep3B and HeLa). All of them expressed lower AhR protein levels, leading to reduced ligand-induced, DRE-driven downstream activity. The AhR protein levels in p23-specific knockdown stable cells were reversed back to wild type levels after exogenous p23 was introduced. Reduction of the AhR protein levels in these stable cells was caused by a decrease in the AhR message levels and an increase of the AhR protein degradation in the absence of ligand. This ligand-independent degradation of AhR was not reversed by MG132, suggesting that the 26S proteasome was not responsible for the degradation. In addition, MG132 could not protect AhR from the ligand-induced degradation in both mouse and human p23-knockdown stable cells.
Copyright © 2012 Elsevier Inc. All rights reserved.
Conflict of interest statement
None.
Figures


















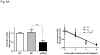
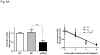
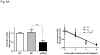
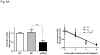

References
-
- Smith DF, Faber LE, Toft DO. Purification of unactivated progesterone receptor and identification of novel receptor-associated proteins. J Biol Chem. 1990;265:3996–4003. - PubMed
-
- Johnson JL, Toft DO. A novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J Biol Chem. 1994;269:24989–93. - PubMed
-
- Johnson JL, Toft DO. Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol Endocrinol. 1995;9:670–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

