Glycoside hydrolases from a targeted compost metagenome, activity-screening and functional characterization
- PMID: 22759983
- PMCID: PMC3477009
- DOI: 10.1186/1472-6750-12-38
Glycoside hydrolases from a targeted compost metagenome, activity-screening and functional characterization
Abstract
Background: Metagenomics approaches provide access to environmental genetic diversity for biotechnology applications, enabling the discovery of new enzymes and pathways for numerous catalytic processes. Discovery of new glycoside hydrolases with improved biocatalytic properties for the efficient conversion of lignocellulosic material to biofuels is a critical challenge in the development of economically viable routes from biomass to fuels and chemicals.
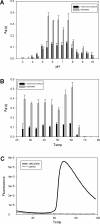
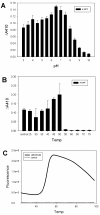
Results: Twenty-two putative ORFs (open reading frames) were identified from a switchgrass-adapted compost community based on sequence homology to related gene families. These ORFs were expressed in E. coli and assayed for predicted activities. Seven of the ORFs were demonstrated to encode active enzymes, encompassing five classes of hemicellulases. Four enzymes were over expressed in vivo, purified to homogeneity and subjected to detailed biochemical characterization. Their pH optima ranged between 5.5 - 7.5 and they exhibit moderate thermostability up to ~60-70°C.
Conclusions: Seven active enzymes were identified from this set of ORFs comprising five different hemicellulose activities. These enzymes have been shown to have useful properties, such as moderate thermal stability and broad pH optima, and may serve as the starting points for future protein engineering towards the goal of developing efficient enzyme cocktails for biomass degradation under diverse process conditions.
Figures


Similar articles
-
Ruminal metagenomic libraries as a source of relevant hemicellulolytic enzymes for biofuel production.Microb Biotechnol. 2018 Jul;11(4):781-787. doi: 10.1111/1751-7915.13269. Epub 2018 Apr 17. Microb Biotechnol. 2018. PMID: 29663699 Free PMC article.
-
Microbial carbohydrate active enzyme (CAZyme) genes and diversity from Menagesha Suba natural forest soils of Ethiopia as revealed by shotgun metagenomic sequencing.BMC Microbiol. 2024 Aug 1;24(1):285. doi: 10.1186/s12866-024-03436-9. BMC Microbiol. 2024. PMID: 39090559 Free PMC article.
-
Characterization of a metagenome-derived protease from contaminated agricultural soil microorganisms and its random mutagenesis.Folia Microbiol (Praha). 2017 Nov;62(6):499-508. doi: 10.1007/s12223-017-0522-y. Epub 2017 Apr 5. Folia Microbiol (Praha). 2017. PMID: 28382524
-
Sucrose Phosphorylase and Related Enzymes in Glycoside Hydrolase Family 13: Discovery, Application and Engineering.Int J Mol Sci. 2020 Apr 5;21(7):2526. doi: 10.3390/ijms21072526. Int J Mol Sci. 2020. PMID: 32260541 Free PMC article. Review.
-
Lignocellulolytic systems of soil bacteria: A vast and diverse toolbox for biotechnological conversion processes.Biotechnol Adv. 2019 Nov 1;37(6):107374. doi: 10.1016/j.biotechadv.2019.03.013. Epub 2019 Mar 22. Biotechnol Adv. 2019. PMID: 30910513 Review.
Cited by
-
Revealing the insoluble metasecretome of lignocellulose-degrading microbial communities.Sci Rep. 2017 May 24;7(1):2356. doi: 10.1038/s41598-017-02506-5. Sci Rep. 2017. PMID: 28539641 Free PMC article.
-
Microbial community structure and dynamics in thermophilic composting viewed through metagenomics and metatranscriptomics.Sci Rep. 2016 Dec 12;6:38915. doi: 10.1038/srep38915. Sci Rep. 2016. PMID: 27941956 Free PMC article.
-
Metagenomic Screening for Lipolytic Genes Reveals an Ecology-Clustered Distribution Pattern.Front Microbiol. 2022 Jun 10;13:851969. doi: 10.3389/fmicb.2022.851969. eCollection 2022. Front Microbiol. 2022. PMID: 35756004 Free PMC article.
-
Screening of multimeric β-xylosidases from the gut microbiome of a higher termite, Globitermes brachycerastes.Int J Biol Sci. 2018 Apr 26;14(6):608-615. doi: 10.7150/ijbs.22763. eCollection 2018. Int J Biol Sci. 2018. PMID: 29904275 Free PMC article.
-
Cellulases from Thermophiles Found by Metagenomics.Microorganisms. 2018 Jul 10;6(3):66. doi: 10.3390/microorganisms6030066. Microorganisms. 2018. PMID: 29996513 Free PMC article. Review.

