Molecular typing of Mycobacterium abscessus based on tandem-repeat polymorphism
- PMID: 22760048
- PMCID: PMC3421802
- DOI: 10.1128/JCM.00753-12
Molecular typing of Mycobacterium abscessus based on tandem-repeat polymorphism
Abstract
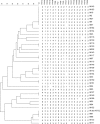
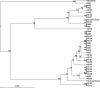
A variable-number tandem-repeat (VNTR) typing assay for the differentiation of Mycobacterium abscessus strains was developed. This assay showed complete reproducibility, locus stability, and a discriminatory power (Hunter-Gaston discriminatory index [HGDI] of 0.9563) that is superior to that of multilocus sequencing. It is a promising tool for the investigation of Mycobacterium abscessus epidemiology and nosocomial outbreaks.
Figures
References
-
- Adékambi T, Berger P, Raoult D, Drancourt M. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133–143 - PubMed
-
- Appelgren P, et al. 2008. Late-onset posttraumatic skin and soft-tissue infections caused by rapid-growing mycobacteria in tsunami survivors. Clin. Infect. Dis. 47:e11–e16 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

