Interhaplotypic heterogeneity and heterochromatic features may contribute to recombination suppression at the S locus in apple (Malusxdomestica)
- PMID: 22760470
- PMCID: PMC3428002
- DOI: 10.1093/jxb/ers176
Interhaplotypic heterogeneity and heterochromatic features may contribute to recombination suppression at the S locus in apple (Malusxdomestica)
Abstract
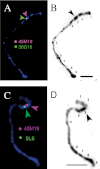
Gametophytic self-incompatibility (GSI) is controlled by a complex S locus containing the pistil determinant S-RNase and pollen determinant SFB/SLF. Tight linkage of the pistil and pollen determinants is necessary to guarantee the self-incompatibility (SI) function. However, multiple probable pollen determinants of apple and Japanese pear, SFBBs (S locus F-box brothers), exist in each S haplotype, and how these multiple genes maintain the SI function remains unclear. It is shown here by high-resolution fluorescence in situ hybridization (FISH) that SFBB genes of the apple S9 haplotype are physically linked to the S9-RNase gene, and the S locus is located in the subtelomeric region. FISH analyses also determined the relative order of SFBB genes and S-RNase in the S9 haplotype, and showed that gene order differs between the S9 and S3 haplotypes. Furthermore, it is shown that the apple S locus is located in a knob-like large heterochromatin block where DNA is highly methylated. It is proposed that interhaplotypic heterogeneity and the heterochromatic nature of the S locus help to suppress recombination at the S locus in apple.
Figures



Similar articles
-
Apple S locus region represents a large cluster of related, polymorphic and pollen-specific F-box genes.Plant Mol Biol. 2010 Sep;74(1-2):143-54. doi: 10.1007/s11103-010-9662-z. Epub 2010 Jul 15. Plant Mol Biol. 2010. PMID: 20628788
-
S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear.Genetics. 2007 Apr;175(4):1869-81. doi: 10.1534/genetics.106.068858. Epub 2007 Jan 21. Genetics. 2007. PMID: 17237509 Free PMC article.
-
Sequence divergence and loss-of-function phenotypes of S locus F-box brothers genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia).Plant J. 2011 Dec;68(6):1028-38. doi: 10.1111/j.1365-313X.2011.04752.x. Epub 2011 Oct 4. Plant J. 2011. PMID: 21851432
-
Self-incompatibility in Petunia: a self/nonself-recognition mechanism employing S-locus F-box proteins and S-RNase to prevent inbreeding.Wiley Interdiscip Rev Dev Biol. 2012 Mar-Apr;1(2):267-75. doi: 10.1002/wdev.10. Epub 2011 Nov 17. Wiley Interdiscip Rev Dev Biol. 2012. PMID: 23801440 Review.
-
Pollen-expressed F-box gene family and mechanism of S-RNase-based gametophytic self-incompatibility (GSI) in Rosaceae.Sex Plant Reprod. 2010 Mar;23(1):39-43. doi: 10.1007/s00497-009-0111-6. Epub 2009 Aug 30. Sex Plant Reprod. 2010. PMID: 20165962 Review.
Cited by
-
Characterization of 25 full-length S-RNase alleles, including flanking regions, from a pool of resequenced apple cultivars.Plant Mol Biol. 2018 Jun;97(3):279-296. doi: 10.1007/s11103-018-0741-x. Epub 2018 May 29. Plant Mol Biol. 2018. PMID: 29845556
-
Isolation and characterization of multiple F-box genes linked to the S9- and S10-RNase in apple (Malus × domestica Borkh.).Plant Reprod. 2013 Jun;26(2):101-11. doi: 10.1007/s00497-013-0212-0. Epub 2013 Feb 12. Plant Reprod. 2013. PMID: 23686223
-
Identification of Putative Markers of Non-infectious Bud Failure in Almond [Prunus dulcis (Mill.) D.A. Webb] Through Genome Wide DNA Methylation Profiling and Gene Expression Analysis in an Almond × Peach Hybrid Population.Front Plant Sci. 2022 Feb 14;13:804145. doi: 10.3389/fpls.2022.804145. eCollection 2022. Front Plant Sci. 2022. PMID: 35237284 Free PMC article.
-
Molecular mechanisms and genetic regulation of self-incompatibility in flowering plants: implications for crop improvement and evolutionary biology.Plant Mol Biol. 2025 Jun 25;115(4):76. doi: 10.1007/s11103-025-01610-9. Plant Mol Biol. 2025. PMID: 40560319 Review.
-
Reciprocal translocation identified in Vigna angularis dominates the wild population in East Japan.J Plant Res. 2015 Jul;128(4):653-63. doi: 10.1007/s10265-015-0720-0. Epub 2015 Mar 22. J Plant Res. 2015. PMID: 25796202
References
-
- Anderson MA, Cornish EC, Mau SL, et al. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata . Nature. 1986;321:38–44.
-
- Bernatzky R. Genetic mapping and protein product diversity of the self-incompatibility locus in wild tomato (Lycopersicon peruvianum) Biochemical Genetics. 1993;31:173–184. - PubMed
-
- Broothaerts W, Janssens GA, Proost P, Broekaert WF. cDNA cloning and molecular analysis of two self-incompatibility alleles from apple. Plant Molecular Biology. 1995;27:499–511. - PubMed

