Conditional gene manipulation: Cre-ating a new biological era
- PMID: 22761243
- PMCID: PMC3390709
- DOI: 10.1631/jzus.B1200042
Conditional gene manipulation: Cre-ating a new biological era
Abstract
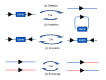
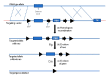
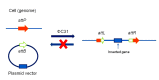
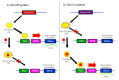
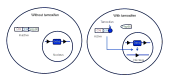
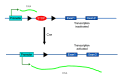
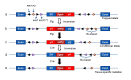
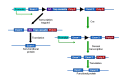
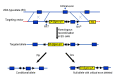
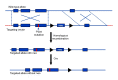
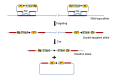
To solve the problem of embryonic lethality in conventional gene knockouts, site-specific recombinase (SSR) systems (Cre-loxP, Flp-FRT, and ΦC31) have been used for tissue-specific gene knockout. With the combination of an SSR system and inducible gene expression systems (tetracycline and tamoxifen), stage-specific knockout and transgenic expression can be achieved. The application of this "SSR+inducible" conditional tool to genomic manipulation can be extended in various ways. Alternatives to conditional gene targeting, such as conditional gene trapping, multipurpose conditional alleles, and conditional gene silencing, have been developed. SSR systems can also be used to construct precise disease models with point mutations and chromosomal abnormalities. With these exciting achievements, we are moving towards a new era in which the whole genome can be manipulated as we wish.
Figures
Similar articles
-
Road to precision: recombinase-based targeting technologies for genome engineering.Curr Opin Biotechnol. 2007 Oct;18(5):411-9. doi: 10.1016/j.copbio.2007.07.013. Epub 2007 Sep 27. Curr Opin Biotechnol. 2007. PMID: 17904350 Review.
-
Strategies for the use of site-specific recombinases in genome engineering.Methods Mol Med. 2005;103:245-57. doi: 10.1385/1-59259-780-7:245. Methods Mol Med. 2005. PMID: 15542911
-
The generation and characterization of novel Col1a1FRT-Cre-ER-T2-FRT and Col1a1FRT-STOP-FRT-Cre-ER-T2 mice for sequential mutagenesis.Dis Model Mech. 2015 Sep;8(9):1155-66. doi: 10.1242/dmm.021204. Epub 2015 Jul 16. Dis Model Mech. 2015. Retraction in: Dis Model Mech. 2016 Dec 1;9(12):1513. doi: 10.1242/dmm.028340. PMID: 26183214 Free PMC article. Retracted.
-
Conditional somatic mutagenesis in the mouse using site-specific recombinases.Handb Exp Pharmacol. 2007;(178):3-28. doi: 10.1007/978-3-540-35109-2_1. Handb Exp Pharmacol. 2007. PMID: 17203649 Review.
-
LoxP-FRT Trap (LOFT): a simple and flexible system for conventional and reversible gene targeting.BMC Biol. 2012 Nov 30;10:96. doi: 10.1186/1741-7007-10-96. BMC Biol. 2012. PMID: 23198860 Free PMC article.
Cited by
-
Tamoxifen affects chronic pancreatitis-related fibrogenesis in an experimental mouse model: an effect beyond Cre recombination.FEBS Open Bio. 2019 Oct;9(10):1756-1768. doi: 10.1002/2211-5463.12714. Epub 2019 Sep 7. FEBS Open Bio. 2019. PMID: 31380604 Free PMC article.
-
An overview of mouse models of hepatocellular carcinoma.Infect Agent Cancer. 2023 Sep 5;18(1):49. doi: 10.1186/s13027-023-00524-9. Infect Agent Cancer. 2023. PMID: 37670307 Free PMC article. Review.
-
CRISPR Cpf1 proteins: structure, function and implications for genome editing.Cell Biosci. 2019 May 9;9:36. doi: 10.1186/s13578-019-0298-7. eCollection 2019. Cell Biosci. 2019. PMID: 31086658 Free PMC article. Review.
-
Animal Models to Study MicroRNA Function.Adv Cancer Res. 2017;135:53-118. doi: 10.1016/bs.acr.2017.06.006. Epub 2017 Aug 8. Adv Cancer Res. 2017. PMID: 28882225 Free PMC article. Review.
-
Discovery of Nigri/nox and Panto/pox site-specific recombinase systems facilitates advanced genome engineering.Sci Rep. 2016 Jul 22;6:30130. doi: 10.1038/srep30130. Sci Rep. 2016. PMID: 27444945 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

