A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity
- PMID: 22761313
- PMCID: PMC3411946
- DOI: 10.1073/pnas.1209134109
A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity
Abstract
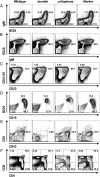
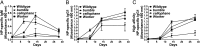
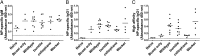
Using chemical germ-line mutagenesis, we screened mice for defects in the humoral immune response to a type II T-independent immunogen and an experimental alphavirus vector. A total of 26 mutations that impair humoral immunity were recovered, and 19 of these mutations have been positionally cloned. Among the phenovariants were bumble, cellophane, and Worker ascribed to mutations in Nfkbid, Zeb1, and Ruvbl2, respectively. We show that IκBNS, the nuclear IκB-like protein encoded by Nfkbid, is required for the development of marginal zone and peritoneal B-1 B cells and additionally required for extrafollicular antibody responses to T-independent and -dependent immunogens. Zeb1 is also required for marginal zone and peritoneal B-1 B-cell development as well as T-cell development, germinal center formation, and memory B-cell responses. Finally, Ruvbl2 is required for T-cell development and maximal T-dependent antibody responses. Collectively, the mutations that we identified give us insight into the points at which disruption of an antibody response can occur. All of the mutations identified to date directly affect lymphocyte development or function; none have an exclusive effect on cells of the innate immune system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

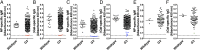


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

