Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1
- PMID: 22761315
- PMCID: PMC3411981
- DOI: 10.1073/pnas.1204991109
Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1
Abstract
Collagenases of the matrix metalloproteinase (MMP) family play major roles in morphogenesis, tissue repair, and human diseases, but how they recognize and cleave the collagen triple helix is not fully understood. Here, we report temperature-dependent binding of a catalytically inactive MMP-1 mutant (E200A) to collagen through the cooperative action of its catalytic and hemopexin domains. Contact between the two molecules was mapped by screening the Collagen Toolkit peptide library and by hydrogen/deuterium exchange. The crystal structure of MMP-1(E200A) bound to a triple-helical collagen peptide revealed extensive interactions of the 115-Å-long triple helix with both MMP-1 domains. An exosite in the hemopexin domain, which binds the leucine 10 residues C-terminal to the scissile bond, is critical for collagenolysis and represents a unique target for inhibitor development. The scissile bond is not correctly positioned for hydrolysis in the crystallized complex. A productive binding mode is readily modeled, without altering the MMP-1 structure or the exosite interactions, by axial rotation of the collagen homotrimer. Interdomain flexing of the enzyme and a localized excursion of the collagen chain closest to the active site, facilitated by thermal loosening of the substrate, may lead to the first transition state of collagenolysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


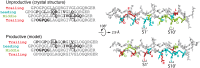

Similar articles
-
Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis.EMBO J. 2004 Aug 4;23(15):3020-30. doi: 10.1038/sj.emboj.7600318. Epub 2004 Jul 15. EMBO J. 2004. PMID: 15257288 Free PMC article.
-
Structural basis for matrix metalloproteinase 1-catalyzed collagenolysis.J Am Chem Soc. 2012 Feb 1;134(4):2100-10. doi: 10.1021/ja208338j. Epub 2012 Jan 19. J Am Chem Soc. 2012. PMID: 22239621 Free PMC article.
-
The interface between catalytic and hemopexin domains in matrix metalloproteinase-1 conceals a collagen binding exosite.J Biol Chem. 2011 Dec 30;286(52):45073-82. doi: 10.1074/jbc.M111.285213. Epub 2011 Oct 26. J Biol Chem. 2011. PMID: 22030392 Free PMC article.
-
Biophysical studies of matrix metalloproteinase/triple-helix complexes.Adv Protein Chem Struct Biol. 2014;97:37-48. doi: 10.1016/bs.apcsb.2014.09.001. Epub 2014 Nov 7. Adv Protein Chem Struct Biol. 2014. PMID: 25458354 Free PMC article. Review.
-
Matrix metalloproteinases and collagen catabolism.Biopolymers. 2002;66(1):19-32. doi: 10.1002/bip.10201. Biopolymers. 2002. PMID: 12228918 Review.
Cited by
-
A synergy between the catalytic and structural Zn(II) ions and the enzyme and substrate dynamics underlies the structure-function relationships of matrix metalloproteinase collagenolysis.J Biol Inorg Chem. 2021 Aug;26(5):583-597. doi: 10.1007/s00775-021-01876-6. Epub 2021 Jul 6. J Biol Inorg Chem. 2021. PMID: 34228191 Free PMC article.
-
Cathepsin K Contributes to Cavitation and Collagen Turnover in Pulmonary Tuberculosis.J Infect Dis. 2016 Feb 15;213(4):618-27. doi: 10.1093/infdis/jiv458. Epub 2015 Sep 27. J Infect Dis. 2016. PMID: 26416658 Free PMC article.
-
Path to Collagenolysis: COLLAGEN V TRIPLE-HELIX MODEL BOUND PRODUCTIVELY AND IN ENCOUNTERS BY MATRIX METALLOPROTEINASE-12.J Biol Chem. 2016 Apr 8;291(15):7888-901. doi: 10.1074/jbc.M115.703124. Epub 2016 Feb 17. J Biol Chem. 2016. PMID: 26887942 Free PMC article.
-
Biochemical characterization and structure determination of a potent, selective antibody inhibitor of human MMP9.J Biol Chem. 2017 Apr 21;292(16):6810-6820. doi: 10.1074/jbc.M116.760579. Epub 2017 Feb 24. J Biol Chem. 2017. PMID: 28235803 Free PMC article.
-
Improved DNA recovery and STR profile development from weathered Bos taurus bones using demineralized bone slices.J Forensic Sci. 2025 May;70(3):954-963. doi: 10.1111/1556-4029.70023. Epub 2025 Mar 16. J Forensic Sci. 2025. PMID: 40090872 Free PMC article.
References
-
- Kadler KE, Baldock C, Bella J, Boot-Handford RP. Collagens at a glance. J Cell Sci. 2007;120:1955–1958. - PubMed
-
- Brodsky B, Persikov AV. Molecular structure of the collagen triple helix. Adv Protein Chem. 2005;70:301–339. - PubMed
-
- Nagase H, Visse R. Triple helicase activity and the structural basis of collagenolysis. In: Parks WC, Mecham RP, editors. Extracellular Matrix Degradation, Biology of Extracellular Matrix. Heidelberg: Springer; 2011. pp. 95–122.
-
- Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: A tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3:207–214. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

