A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny
- PMID: 22761372
- PMCID: PMC3446554
- DOI: 10.1128/JVI.01278-12
A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny
Abstract
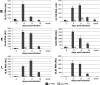
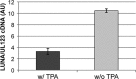
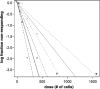
Human cytomegalovirus (HCMV) is a herpesvirus that establishes a lifelong, latent infection within a host. At times when the immune system is compromised, the virus undergoes a lytic reactivation producing infectious progeny. The identification and understanding of the biological mechanisms underlying HCMV latency and reactivation are not completely defined. To this end, we have developed a tractable in vitro model system to investigate these phases of viral infection using a clonal population of myeloid progenitor cells (Kasumi-3 cells). Infection of these cells results in maintenance of the viral genome with restricted viral RNA expression that is reversed with the addition of the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA, also known as PMA). Additionally, a latent viral transcript (LUNA) is expressed at times where viral lytic transcription is suppressed. Infected Kasumi-3 cells initiate production of infectious virus following TPA treatment, which requires cell-to-cell contact for efficient transfer of virus to other cell types. Importantly, lytically infected fibroblast, endothelial, or epithelial cells can transfer virus to Kasumi-3 cells, which fail to initiate lytic replication until stimulated with TPA. Finally, inflammatory cytokines, in addition to the pharmacological agent TPA, are sufficient for transcription of immediate-early (IE) genes following latent infection. Taken together, our findings argue that the Kasumi-3 cell line is a tractable in vitro model system with which to study HCMV latency and reactivation.
Figures






References
-
- Alain S, et al. 1993. Rapid detection of cytomegalovirus strains resistant to ganciclovir through mutations within the gene UL97. Mol. Cell. Probes 7:487–495 - PubMed
-
- Andrews PW, et al. 1984. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab. Invest. 50:147–162 - PubMed
-
- Bego MG, Keyes LR, Maciejewski J, St Jeor SC. 2011. Human cytomegalovirus latency-associated protein LUNA is expressed during HCMV infections in vivo. Arch. Virol. 156:1847–1851 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

