Kaposi's sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIβ for latent DNA replication of the terminal repeats
- PMID: 22761383
- PMCID: PMC3446556
- DOI: 10.1128/JVI.00839-12
Kaposi's sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIβ for latent DNA replication of the terminal repeats
Abstract
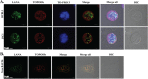
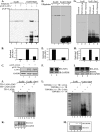
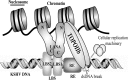
The latency-associated nuclear antigen (LANA) encoded by Kaposi's sarcoma-associated herpesvirus (KSHV) plays a major role in maintaining latency and is critical for the perpetual segregation of viral episomes to the progeny nuclei of newly divided cells. LANA binds to KSHV terminal repeat (TR) DNA and tethers the viral episomes to host chromosomes through the association of chromatin-bound cellular proteins. TR elements serve as potential origin sites of KSHV replication and have been shown to play important roles in latent DNA replication and transcription of adjacent genes. Affinity chromatography and proteomics analysis using KSHV TR DNA and the LANA binding site as the affinity column identified topoisomerase IIβ (TopoIIβ) as a LANA-interacting protein. Here, we show that TopoIIβ forms complexes with LANA that colocalize as punctuate bodies in the nucleus of KSHV-infected cells. The specific TopoIIβ binding region of LANA has been identified to its N terminus and the first 32 amino acid residues containing the nucleosome-binding region crucial for binding. Moreover, this region could also act as a dominant negative to disrupt association of TopoIIβ with LANA. TopoIIβ plays an important role in LANA-dependent latent DNA replication, as addition of ellipticine, a selective inhibitor of TopoII, negatively regulated replication mediated by the TR. DNA break labeling and chromatin immunoprecipitation assay using biotin-16-dUTP and terminal deoxynucleotide transferase showed that TopoIIβ mediates a transient DNA break on viral DNA. These studies confirm that LANA recruits TopoIIβ at the origins of latent replication to unwind the DNA for replication.
Figures





References
-
- An FQ, et al. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280:3862–3874 - PubMed
-
- Ballestas ME, Chatis PA, Kaye KM. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641–644 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

