Hfq proximity and orientation controls RNA annealing
- PMID: 22761405
- PMCID: PMC3458560
- DOI: 10.1093/nar/gks618
Hfq proximity and orientation controls RNA annealing
Abstract
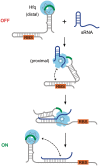
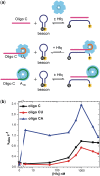
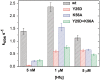
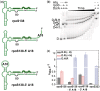
Regulation of bacterial gene networks by small non-coding RNAs (sRNAs) requires base pairing with messenger RNA (mRNA) targets, which is facilitated by Hfq protein. Hfq is recruited to sRNAs and mRNAs through U-rich- and A-rich-binding sites, respectively, but their distance from the sRNA-mRNA complementary region varies widely among different genes. To determine whether distance and binding orientation affect Hfq's chaperone function, we engineered 'toy' RNAs containing strong Hfq-binding sites at defined distances from the complementary target site. We show that RNA annealing is fastest when the distal face of Hfq binds an A-rich sequence immediately 3' of the target. This recruitment advantage is lost when Hfq binds >20 nt away from the target, but is partially restored by secondary structure that shortens this distance. Although recruitment through Hfq's distal face accelerates RNA annealing, tight binding of six Us to Hfq's proximal face inhibits annealing. Finally, we show that ectopic A-rich motifs dramatically accelerate base pairing between DsrA sRNA and a minimal rpoS mRNA in the presence of Hfq, demonstrating that proximity and orientation predict the activity of Hfq on long RNAs.
Figures


References
-
- Kaberdin VR, Blasi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 2006;30:967–979. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

