Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion
- PMID: 22761418
- PMCID: PMC3436143
- DOI: 10.1074/jbc.M112.367862
Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion
Abstract
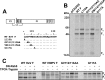
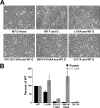
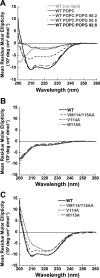
Viral fusion proteins are intriguing molecular machines that undergo drastic conformational changes to facilitate virus-cell membrane fusion. During fusion a hydrophobic region of the protein, termed the fusion peptide (FP), is inserted into the target host cell membrane, with subsequent conformational changes culminating in membrane merger. Class I fusion proteins contain FPs between 20 and 30 amino acids in length that are highly conserved within viral families but not between. To examine the sequence dependence of the Hendra virus (HeV) fusion (F) protein FP, the first eight amino acids were mutated first as double, then single, alanine mutants. Mutation of highly conserved glycine residues resulted in inefficient F protein expression and processing, whereas substitution of valine residues resulted in hypofusogenic F proteins despite wild-type surface expression levels. Synthetic peptides corresponding to a portion of the HeV F FP were shown to adopt an α-helical secondary structure in dodecylphosphocholine micelles and small unilamellar vesicles using circular dichroism spectroscopy. Interestingly, peptides containing point mutations that promote lower levels of cell-cell fusion within the context of the whole F protein were less α-helical and induced less membrane disorder in model membranes. These data represent the first extensive structure-function relationship of any paramyxovirus FP and demonstrate that the HeV F FP and potentially other paramyxovirus FPs likely require an α-helical structure for efficient membrane disordering and fusion.
Figures

Similar articles
-
Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state.J Virol. 2004 Dec;78(24):13727-42. doi: 10.1128/JVI.78.24.13727-13742.2004. J Virol. 2004. PMID: 15564482 Free PMC article.
-
Transmembrane Domain Dissociation Is Required for Hendra Virus F Protein Fusogenic Activity.J Virol. 2019 Oct 29;93(22):e01069-19. doi: 10.1128/JVI.01069-19. Print 2019 Nov 15. J Virol. 2019. PMID: 31462574 Free PMC article.
-
Third Helical Domain of the Nipah Virus Fusion Glycoprotein Modulates both Early and Late Steps in the Membrane Fusion Cascade.J Virol. 2020 Sep 15;94(19):e00644-20. doi: 10.1128/JVI.00644-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32669342 Free PMC article.
-
Structural and functional specificity of Influenza virus haemagglutinin and paramyxovirus fusion protein anchoring peptides.Virus Res. 2017 Jan 2;227:183-199. doi: 10.1016/j.virusres.2016.09.014. Epub 2016 Oct 20. Virus Res. 2017. PMID: 27773768 Review.
-
Activation of paramyxovirus membrane fusion and virus entry.Curr Opin Virol. 2014 Apr;5:24-33. doi: 10.1016/j.coviro.2014.01.005. Epub 2014 Feb 16. Curr Opin Virol. 2014. PMID: 24530984 Free PMC article. Review.
Cited by
-
The three lives of viral fusion peptides.Chem Phys Lipids. 2014 Jul;181:40-55. doi: 10.1016/j.chemphyslip.2014.03.003. Epub 2014 Apr 2. Chem Phys Lipids. 2014. PMID: 24704587 Free PMC article. Review.
-
Conformation and lipid interaction of the fusion peptide of the paramyxovirus PIV5 in anionic and negative-curvature membranes from solid-state NMR.J Am Chem Soc. 2014 Feb 12;136(6):2611-24. doi: 10.1021/ja4121956. Epub 2014 Jan 30. J Am Chem Soc. 2014. PMID: 24428385 Free PMC article.
-
Unity in diversity: shared mechanism of entry among paramyxoviruses.Prog Mol Biol Transl Sci. 2015;129:1-32. doi: 10.1016/bs.pmbts.2014.10.001. Epub 2014 Dec 1. Prog Mol Biol Transl Sci. 2015. PMID: 25595799 Free PMC article. Review.
-
Parainfluenza virus 5 fusion protein maintains pre-fusion stability but not fusogenic activity following mutation of a transmembrane leucine/isoleucine domain.J Gen Virol. 2020 May;101(5):467-472. doi: 10.1099/jgv.0.001399. Epub 2020 Feb 25. J Gen Virol. 2020. PMID: 32100701 Free PMC article.
-
A Hydrophobic Target: Using the Paramyxovirus Fusion Protein Transmembrane Domain To Modulate Fusion Protein Stability.J Virol. 2019 Aug 13;93(17):e00863-19. doi: 10.1128/JVI.00863-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31217248 Free PMC article.
References
-
- Lamb R. A., Parks G. D. (2007) in Fields Virology (Knipe D. M., Howley P. M., eds.) pp 1449–1496, Fifth Ed., Lippincott, Williams and Wilkins, Philadelphia
-
- O'Sullivan J. D., Allworth A. M., Paterson D. L., Snow T. M., Boots R., Gleeson L. J., Gould A. R., Hyatt A. D., Bradfield J. (1997) Fatal encephalitis due to novel Paramyxovirus transmitted from horses. Lancet 349, 93–95 - PubMed
-
- Harcourt B. H., Tamin A., Ksiazek T. G., Rollin P. E., Anderson L. J., Bellini W. J., Rota P. A. (2000) Molecular characterization of Nipah virus, a newly emergent paramxyovirus. Virology 271, 334–349 - PubMed
-
- Murray K., Selleck P., Hooper P., Hyatt A., Gould A., Gleeson L., Westbury H., Hiley L., Selvey L., Rodwell B. (1995) A morbillivirus that caused fatal disease in horses and humans. Science 268, 94–97 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

