Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1
- PMID: 22761421
- PMCID: PMC3436169
- DOI: 10.1074/jbc.M112.378281
Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1
Abstract
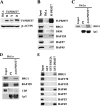
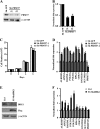
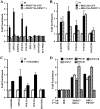
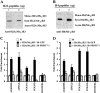
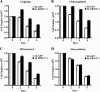
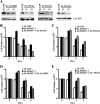
Covalent modification of histones by protein arginine methyltransferases (PRMTs) impacts genome organization and gene expression. In this report, we show that PRMT7 interacts with the BRG1-based hSWI/SNF chromatin remodeling complex and specifically methylates histone H2A Arg-3 (H2AR3) and histone H4 Arg-3 (H4R3). To elucidate the biological function of PRMT7, we knocked down its expression in NIH 3T3 cells and analyzed global gene expression. Our findings show that PRMT7 negatively regulates expression of genes involved in DNA repair, including ALKBH5, APEX2, POLD1, and POLD2. Chromatin immunoprecipitation (ChIP) revealed that PRMT7 and dimethylated H2AR3 and H4R3 are enriched at target DNA repair genes in parental cells, whereas PRMT7 knockdown caused a significant decrease in PRMT7 recruitment and H2AR3/H4R3 methylation. Decreased PRMT7 expression also resulted in derepression of target DNA repair genes and enhanced cell resistance to DNA-damaging agents. Furthermore, we show that BRG1 co-localizes with PRMT7 on target promoters and that expression of a catalytically inactive form of BRG1 results in derepression of PRMT7 target DNA repair genes. Remarkably, reducing expression of individual PRMT7 target DNA repair genes showed that only the catalytic subunit of DNA polymerase, POLD1, was able to resensitize PRMT7 knock-down cells to DNA-damaging agents. These results provide evidence for the important role played by PRMT7 in epigenetic regulation of DNA repair genes and cellular response to DNA damage.
Figures

References
-
- Henikoff S. (2008) Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9, 15–26 - PubMed
-
- Klose R. J., Zhang Y. (2007) Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8, 307–318 - PubMed
-
- Lee K. K., Workman J. L. (2007) Histone acetyltransferase complexes. One size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

