Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer
- PMID: 22761427
- PMCID: PMC3436145
- DOI: 10.1074/jbc.M112.366518
Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer
Abstract
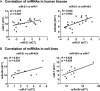
MicroRNAs (miRNAs) are small non-coding RNAs that posttranscriptionally regulate gene expression during many biological processes. Recently, the aberrant expressions of miRNAs have become a major focus in cancer research. The purpose of this study was to identify deregulated miRNAs in oral cancer and further focus on specific miRNAs that were related to patient survival. Here, we report that miRNA expression profiling provided more precise information when oral squamous cell carcinomas were subcategorized on the basis of clinicopathological parameters (tumor primary site, histological subtype, tumor stage, and HPV16 status). An innovative radar chart analysis method was developed to depict subcategories of cancers taking into consideration the expression patterns of multiple miRNAs combined with the clinicopathological parameters. Keratinization of tumors and the high expression of miR-21 were the major factors related to the poor prognosis of patients. Interestingly, a majority of the keratinized tumors expressed high levels of miR-21. Further investigations demonstrated the regulation of the tumor suppressor gene reversion-inducing cysteine-rich protein with kazal motifs (RECK) by two keratinization-associated miRNAs, miR-7 and miR-21. Transfection of miR-7 and miR-21-mimics reduced the expression of RECK through direct miRNA-mediated regulation, and these miRNAs were inversely correlated with RECK in CAL 27 orthotopic xenograft tumors. Furthermore, a similar inverse correlation was demonstrated in CAL 27 cells treated in vitro by different external stimuli such as trypsinization, cell density, and serum concentration. Taken together, our data show that keratinization is associated with poor prognosis of oral cancer patients and keratinization-associated miRNAs mediate deregulation of RECK which may contribute to the aggressiveness of tumors.
Figures






References
-
- Scully C., Felix D. H. (2006) Oral medicine. Update for the dental practitioner oral cancer. Br. Dent. J. 200, 13–17 - PubMed
-
- Jemal A., Tiwari R. C., Murray T., Ghafoor A., Samuels A., Ward E., Feuer E. J., Thun M. J. (2004) Cancer statistics, 2004. CA. Cancer J. Clin. 54, 8–29 - PubMed
-
- Lin W. H., Chen I. H., Wei F. C., Huang J. J., Kang C. J., Hsieh L. L., Wang H. M., Huang S. F. (2011) Clinical significance of preoperative squamous cell carcinoma antigen in oral-cavity squamous cell carcinoma. Laryngoscope 121, 971–977 - PubMed
-
- Méndez E., Houck J. R., Doody D. R., Fan W., Lohavanichbutr P., Rue T. C., Yueh B., Futran N. D., Upton M. P., Farwell D. G., Heagerty P. J., Zhao L. P., Schwartz S. M., Chen C. (2009) A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin. Cancer Res. 15, 1353–1361 - PMC - PubMed
-
- Chung C. H., Parker J. S., Karaca G., Wu J., Funkhouser W. K., Moore D., Butterfoss D., Xiang D., Zanation A., Yin X., Shockley W. W., Weissler M. C., Dressler L. G., Shores C. G., Yarbrough W. G., Perou C. M. (2004) Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 5, 489–500 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

