Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions
- PMID: 22761572
- PMCID: PMC3386243
- DOI: 10.1371/journal.ppat.1002761
Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions
Abstract
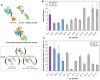
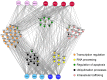
Human Papillomaviruses (HPV) cause widespread infections in humans, resulting in latent infections or diseases ranging from benign hyperplasia to cancers. HPV-induced pathologies result from complex interplays between viral proteins and the host proteome. Given the major public health concern due to HPV-associated cancers, most studies have focused on the early proteins expressed by HPV genotypes with high oncogenic potential (designated high-risk HPV or HR-HPV). To advance the global understanding of HPV pathogenesis, we mapped the virus/host interaction networks of the E2 regulatory protein from 12 genotypes representative of the range of HPV pathogenicity. Large-scale identification of E2-interaction partners was performed by yeast two-hybrid screenings of a HaCaT cDNA library. Based on a high-confidence scoring scheme, a subset of these partners was then validated for pair-wise interaction in mammalian cells with the whole range of the 12 E2 proteins, allowing a comparative interaction analysis. Hierarchical clustering of E2-host interaction profiles mostly recapitulated HPV phylogeny and provides clues to the involvement of E2 in HPV infection. A set of cellular proteins could thus be identified discriminating, among the mucosal HPV, E2 proteins of HR-HPV 16 or 18 from the non-oncogenic genital HPV. The study of the interaction networks revealed a preferential hijacking of highly connected cellular proteins and the targeting of several functional families. These include transcription regulation, regulation of apoptosis, RNA processing, ubiquitination and intracellular trafficking. The present work provides an overview of E2 biological functions across multiple HPV genotypes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Comment in
-
Human papillomavirus E2 protein: a potential key regulator of viral cell pathogenesis.Pathog Glob Health. 2012 Jul;106(3):141. doi: 10.1179/204777312X13462106637765. Pathog Glob Health. 2012. PMID: 23265369 Free PMC article. No abstract available.
Similar articles
-
Control of viral replication and transcription by the papillomavirus E8^E2 protein.Virus Res. 2017 Mar 2;231:96-102. doi: 10.1016/j.virusres.2016.11.005. Epub 2016 Nov 4. Virus Res. 2017. PMID: 27825778 Review.
-
[Virological and carcinogenic aspects of HPV].Bull Acad Natl Med. 2007 Mar;191(3):611-23; discussion 623. Bull Acad Natl Med. 2007. PMID: 18072657 Review. French.
-
Proteomic approaches to the study of papillomavirus-host interactions.Virology. 2013 Jan 5;435(1):57-69. doi: 10.1016/j.virol.2012.09.046. Virology. 2013. PMID: 23217616 Free PMC article. Review.
-
The SMC5/6 Complex Interacts with the Papillomavirus E2 Protein and Influences Maintenance of Viral Episomal DNA.J Virol. 2018 Jul 17;92(15):e00356-18. doi: 10.1128/JVI.00356-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29848583 Free PMC article.
-
Comparative analysis of virus-host interactomes with a mammalian high-throughput protein complementation assay based on Gaussia princeps luciferase.Methods. 2012 Dec;58(4):349-59. doi: 10.1016/j.ymeth.2012.07.029. Epub 2012 Aug 8. Methods. 2012. PMID: 22898364 Free PMC article.
Cited by
-
A comparative approach to characterize the landscape of host-pathogen protein-protein interactions.J Vis Exp. 2013 Jul 18;(77):e50404. doi: 10.3791/50404. J Vis Exp. 2013. PMID: 23893119 Free PMC article.
-
Human papillomavirus E2 protein: a potential key regulator of viral cell pathogenesis.Pathog Glob Health. 2012 Jul;106(3):141. doi: 10.1179/204777312X13462106637765. Pathog Glob Health. 2012. PMID: 23265369 Free PMC article. No abstract available.
-
Inferring Virus-Host relationship between HPV and its host Homo sapiens using protein interaction network.Sci Rep. 2020 May 26;10(1):8719. doi: 10.1038/s41598-020-65837-w. Sci Rep. 2020. PMID: 32457456 Free PMC article.
-
Pyk2 Regulates Human Papillomavirus Replication by Tyrosine Phosphorylation of the E2 Protein.J Virol. 2020 Sep 29;94(20):e01110-20. doi: 10.1128/JVI.01110-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32727877 Free PMC article.
-
CCHCR1 interacts specifically with the E2 protein of human papillomavirus type 16 on a surface overlapping BRD4 binding.PLoS One. 2014 Mar 24;9(3):e92581. doi: 10.1371/journal.pone.0092581. eCollection 2014. PLoS One. 2014. PMID: 24664238 Free PMC article.
References
-
- zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–265. - PubMed
-
- Biliris KA, Koumantakis E, Dokianakis DN, Sourvinos G, Spandidos DA. Human papillomavirus infection of non-melanoma skin cancers in immunocompetent hosts. Cancer Lett. 2000;161:83–88. - PubMed
-
- Wiley D, Masongsong E. Human papillomavirus: the burden of infection. Obstet Gynecol Surv. 2006;61:S3–14. - PubMed
-
- Desaintes C, Demeret C. Control of papillomavirus DNA replication and transcription. Semin Cancer Biol. 1996;7:339–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

